Allergy Asthma Immunol Res.
2017 May;9(3):247-256. 10.4168/aair.2017.9.3.247.
Investigation of the Possible Role of the Hippo/YAP1 Pathway in Asthma and Allergy
- Affiliations
-
- 1Department of Genetics, Cell- and Immunobiology, Semmelweis University, Budapest, Hungary. szalaics@gmail.com
- 2Heim, Pal Children Hospital, Budapest, Hungary.
- 3Department of Pulmonology, Semmelweis University, Budapest, Hungary.
- 4Department of Measurement and Information Systems, University of Technology and Economics, Budapest, Hungary.
- KMID: 2371725
- DOI: http://doi.org/10.4168/aair.2017.9.3.247
Abstract
- PURPOSE
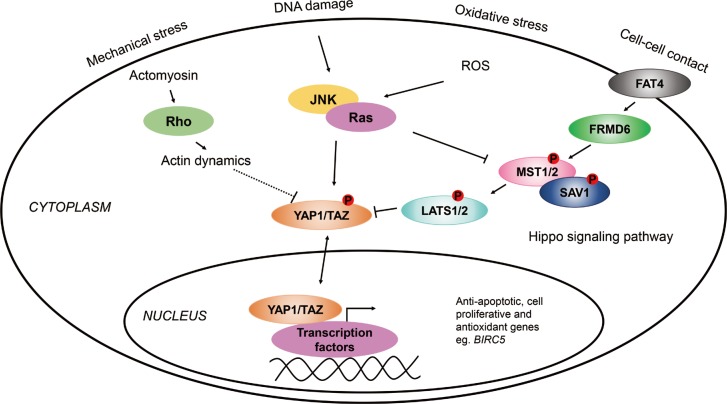
Several lines of evidence indicate that the Hippo/Yes-associated protein 1 (YAP1) pathways might play a role in the pathogenesis of asthma. To investigate the possible role of the Hippo/YAP1 pathway in the pathogenesis of asthma or its phenotypes.
METHODS
The levels of gene expressions of the members of the Hippo/YAP1 were compared. The presence of the proteins of the YAP1 and FRMD6 were analyzed with Western blot in induced sputum of 18 asthmatic subjects and 10 control subjects. Fourteen single nucleotide polymorphisms (SNPs) in the YAP1 gene were genotyped in 522 asthmatic subjects and 711 healthy controls. The results were evaluated with traditional frequentist methods and with Bayesian network-based Bayesian multilevel analysis of relevance (BN-BMLA).
RESULTS
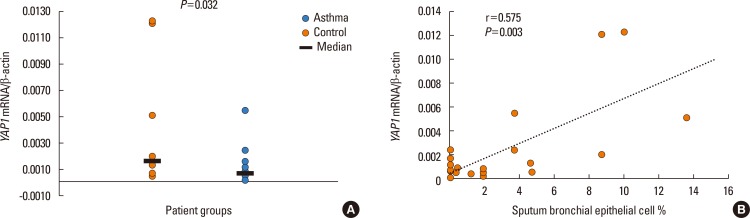
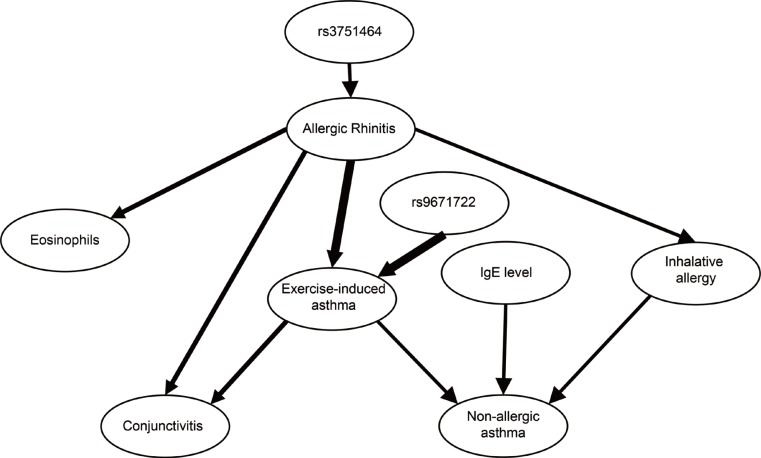
The mRNA of all the members of the Hippo/YAP1 pathway could be detected in the induced sputum of both controls and cases. A correlation was found between YAP1 mRNA levels and sputum bronchial epithelial cells (r=0.575, P=0.003). The signal for the FRMD6 protein could be detected in all sputum samples while the YAP1 protein could not be detected in the sputum samples, of the healthy controls and severe asthmatics, but it was detectable in mild asthmatics. The rs2846836 SNP of the YAP1 gene was significantly associated with exercise-induced asthma (odds ratio [OR]=2.1 [1.3-3.4]; P=0.004). The distribution of genotypes of rs11225138 and certain haplotypes of the YAP1 gene showed significant differences between different asthma severity statuses. With BN-BMLA, 2 SNPs, genetic variations in the FRMD6 gene proved to be the most relevant to exercise-induced asthma and allergic rhinitis. These 2 SNPs through allergic rhinitis and exercise-induced asthma were in epistatic interaction with each other.
CONCLUSIONS
Our results provided additional evidence that the FRMD6/Hippo/YAP1 pathway plays a role in the pathogenesis of asthma. If additional studies can confirm these findings, this pathway can be a potential novel therapeutic target in asthma and other inflammatory airway diseases.
MeSH Terms
Figure
Reference
-
1. Chen Y, Wong GW, Li J. Environmental exposure and genetic predisposition as risk factors for asthma in China. Allergy Asthma Immunol Res. 2016; 8:92–100. PMID: 26739401.
Article2. Yoon HI. Respiratory review of 2014: asthma. Tuberc Respir Dis. 2014; 77:237–242.
Article3. Meyers DA, Bleecker ER, Holloway JW, Holgate ST. Asthma genetics and personalised medicine. Lancet Respir Med. 2014; 2:405–415. PMID: 24794577.4. Vahedi G, Richard AC, O'Shea JJ. Enhancing the understanding of asthma. Nat Immunol. 2014; 15:701–703. PMID: 25045871.
Article5. Tizaoui K, Kaabachi W, Hamzaoui K, Hamzaoui A. Association of single nucleotide polymorphisms in toll-like receptor genes with asthma risk: a systematic review and meta-analysis. Allergy Asthma Immunol Res. 2015; 7:130–140. PMID: 25729620.
Article6. Zhao CN, Fan Y, Huang JJ, Zhang HX, Gao T, Wang C, et al. The association of GSDMB and ORMDL3 gene polymorphisms with asthma: a meta-analysis. Allergy Asthma Immunol Res. 2015; 7:175–185. PMID: 25729625.7. Huang J, Wu S, Barrera J, Matthews K, Pan D. The hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005; 122:421–434. PMID: 16096061.
Article8. Angus L, Moleirinho S, Herron L, Sinha A, Zhang X, Niestrata M, et al. Willin/FRMD6 expression activates the hippo signaling pathway kinases in mammals and antagonizes oncogenic YAP. Oncogene. 2012; 31:238–250. PMID: 21666719.
Article9. Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, et al. Inactivation of YAP oncoprotein by the hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007; 21:2747–2761. PMID: 17974916.
Article10. Alarcón C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGFβ pathways. Cell. 2009; 139:757–769. PMID: 19914168.
Article11. Strano S, Munarriz E, Rossi M, Castagnoli L, Shaul Y, Sacchi A, et al. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J Biol Chem. 2001; 276:15164–15173. PMID: 11278685.
Article12. Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001; 15:1229–1241. PMID: 11358867.
Article13. Mahoney JE, Mori M, Szymaniak AD, Varelas X, Cardoso WV. The hippo pathway effector Yap controls patterning and differentiation of airway epithelial progenitors. Dev Cell. 2014; 30:137–150. PMID: 25043473.
Article14. Lange AW, Sridharan A, Xu Y, Stripp BR, Perl AK, Whitsett JA. Hippo/Yap signaling controls epithelial progenitor cell proliferation and differentiation in the embryonic and adult lung. J Mol Cell Biol. 2015; 7:35–47. PMID: 25480985.
Article15. Ungvári I, Hullám G, Antal P, Kiszel PS, Gézsi A, Hadadi É, et al. Evaluation of a partial genome screening of two asthma susceptibility regions using bayesian network based bayesian multilevel analysis of relevance. PLoS One. 2012; 7:e33573. PMID: 22432035.
Article16. Ungvári I, Hadadi E, Virág V, Bikov A, Nagy A, Semsei ÁF, et al. Implication of BIRC5 in asthma pathogenesis. Int Immunol. 2012; 24:293–301. PMID: 22336533.17. Wenzel SE. Asthma: defining of the persistent adult phenotypes. Lancet. 2006; 368:804–813. PMID: 16935691.
Article18. Antal P, Millinghoffer A, Hullám G, Hajós G, Sárközy P, Gézsi A, et al. Bayesian, systems-based, multilevel analysis of associations for complex phenotypes: from interpretation to decisions. In : Sinoquet C, Mourad R, editors. Probabilistic graphical models for genetics, genomics, and postgenomics. Oxford: Oxford University Press;2014. p. 318–362.19. Antal P, Millinghoffer A, Hullám G, Hajós G, Szalai C, Falus A. A bioinformatic platform for a bayesian, multiphased, multilevel analysis in immunogenomics. In : Flower DR, Davies M, Ranganathan S, editors. Bioinformatics for immunomics. New York (NY): Springer;2010. p. 157–185.20. Hullám G, Antal P, Szalai C, Falus A. Evaluation of a Bayesian model-based approach in GA studies. JMLR Workshop Conf Proc. 2010; 8:30–43.21. Antal P, Millinghoffer A, Hullám G, Szalai C, Falus AA. Bayesian view of challenges in feature selection: multilevel analysis, feature aggregation, multiple targets, redundancy and interaction. JMLR Workshop Conf Proc. 2008; 4:74–89.22. Gézsi A, Lautner-Csorba O, Erdélyi DJ, Hullám G, Antal P, Semsei ÁF, et al. In interaction with gender a common CYP3A4 polymorphism may influence the survival rate of chemotherapy for childhood acute lymphoblastic leukemia. Pharmacogenomics J. 2015; 15:241–247. PMID: 25266680.
Article23. Lautner-Csorba O, Gézsi A, Erdélyi DJ, Hullám G, Antal P, Semsei ÁF, et al. Roles of genetic polymorphisms in the folate pathway in childhood acute lymphoblastic leukemia evaluated by Bayesian relevance and effect size analysis. PLoS One. 2013; 8:e69843. PMID: 23940529.
Article24. Lautner-Csorba O, Gézsi A, Semsei AF, Antal P, Erdélyi DJ, Schermann G, et al. Candidate gene association study in pediatric acute lymphoblastic leukemia evaluated by Bayesian network based Bayesian multilevel analysis of relevance. BMC Med Genomics. 2012; 5:42. PMID: 23021489.
Article25. Chan TK, Loh XY, Peh HY, Tan WN, Tan WS, Li N, et al. House dust mite-induced asthma causes oxidative damage and DNA double-strand breaks in the lungs. J Allergy Clin Immunol. 2016; 138:84–96. e1PMID: 27157131.
Article26. Mao B, Gao Y, Bai Y, Yuan Z. Hippo signaling in stress response and homeostasis maintenance. Acta Biochim Biophys Sin (Shanghai). 2015; 47:2–9. PMID: 25476206.
Article27. Piccolo S, Dupont S, Cordenonsi M. The biology of YAP/TAZ: hippo signaling and beyond. Physiol Rev. 2014; 94:1287–1312. PMID: 25287865.
Article28. Yin MX, Zhang L. Hippo signaling in epithelial stem cells. Acta Biochim Biophys Sin (Shanghai). 2015; 47:39–45. PMID: 25476205.
Article29. Moleirinho S, Patrick C, Tilston-Lünel AM, Higginson JR, Angus L, Antkowiak M, et al. Willin, an upstream component of the hippo signaling pathway, orchestrates mammalian peripheral nerve fibroblasts. PLoS One. 2013; 8:e60028. PMID: 23593160.
Article30. Jeffery PK, Wardlaw AJ, Nelson FC, Collins JV, Kay AB. Bronchial biopsies in asthma. An ultrastructural, quantitative study and correlation with hyperreactivity. Am Rev Respir Dis. 1989; 140:1745–1753. PMID: 2690708.
Article31. Beasley R, Roche WR, Roberts JA, Holgate ST. Cellular events in the bronchi in mild asthma and after bronchial provocation. Am Rev Respir Dis. 1989; 139:806–817. PMID: 2923380.
Article32. Laitinen LA, Heino M, Laitinen A, Kava T, Haahtela T. Damage of the airway epithelium and bronchial reactivity in patients with asthma. Am Rev Respir Dis. 1985; 131:599–606. PMID: 3994155.
Article