Nat Prod Sci.
2016 Sep;22(3):220-224. 10.20307/nps.2016.22.3.220.
Anti-Helicobacter pylori Compounds from Polygonum cuspidatum
- Affiliations
-
- 1College of Pharmacy and Research Institute of Pharmaceutical Sciences, Gyeongsang National University, Jinju 52828, Korea. amj5812@gnu.ac.kr
- KMID: 2355149
- DOI: http://doi.org/10.20307/nps.2016.22.3.220
Abstract
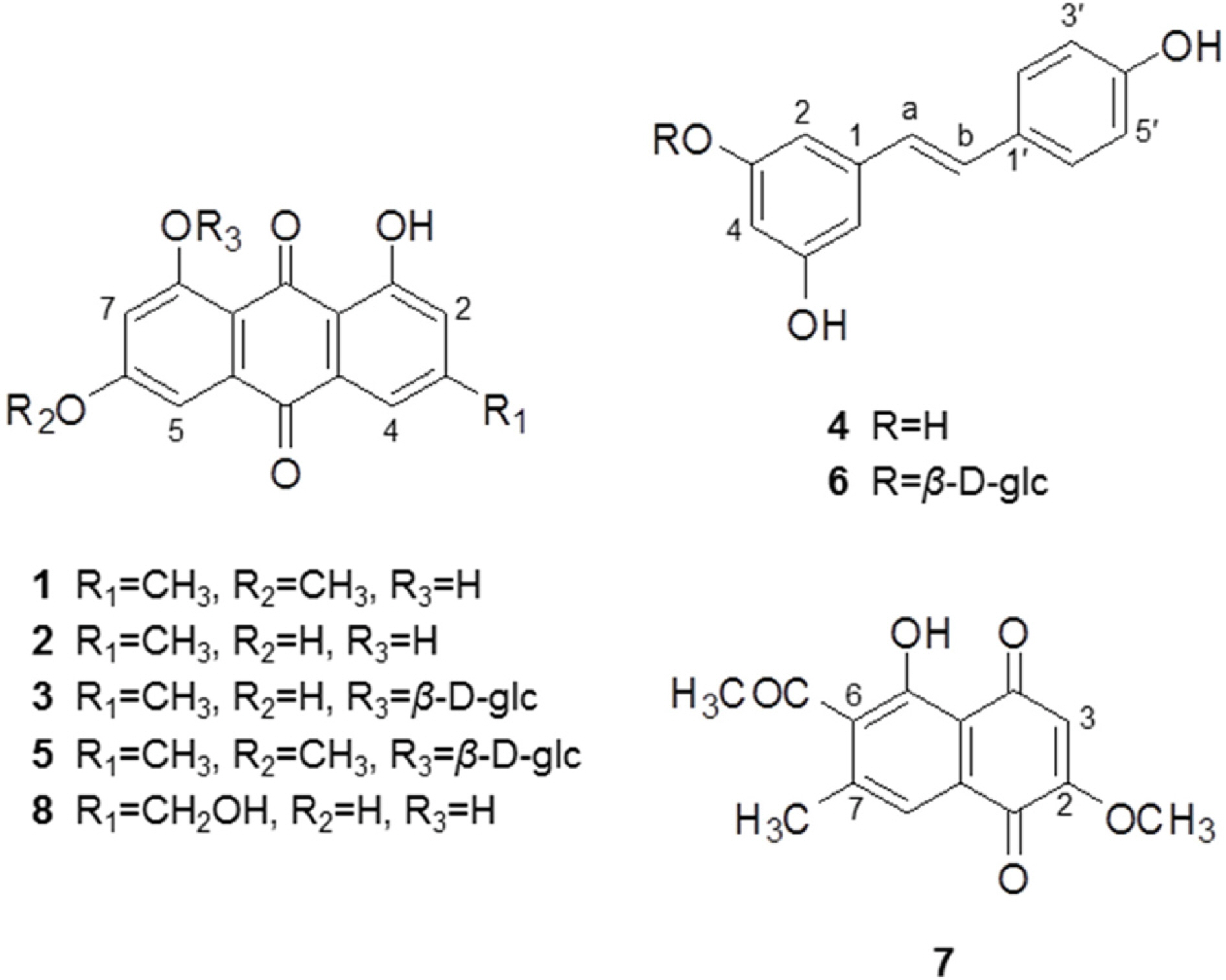
- Anti-Helicobacter pylori activity guided fractionation led to the isolation of five anthraquinones, two stilbenes and one naphthoquinone from the EtOAc fraction of Polygonum cuspidatum, using silica gel column chromatography, Sephadex-LH20, MPLC and recrystallization. The chemical structures were identified to be physcion (1), emodin (2), anthraglycoside B (3), trans-resveratrol (4), anthraglycoside A (5), polydatin (6), 2-methoxy-6-acetyl-7-methyljuglone (7) and citreorosein (8) by UV, ¹H-NMR, ¹³C-NMR and mass spectrometry. Anti-Helicobacter pylori activity including MIC values of each compound was evaluated. All of the isolates exhibited anti-H. pylori activity of which MIC values were lower than that of a positive control, quercetin. Compounds 2 and 7 showed potent growth inhibitory activity. Especially, a naphthoquinone, compound 7 displayed most potent antibacterial activity with MICâ‚…â‚€ value of 0.30 µM and MIC₉₀ value of 0.39 µM. Although anti-H. pylori activity of this plant was previously reported, this is the first report on that of compounds isolated from this species. From these findings, P. cuspidatum roots or its isolates may be useful for H. pylori infection and further study is needed to elucidate mechanism of action.
Keyword
MeSH Terms
Figure
Reference
-
(1). Goodwin C. S., Armstrong J. A., Chilvers T., Peters M., Collins M. D., Sly L., McConnell W., Harper W. E. S.Int. J. Syst. Bacteriol. 1989; 39:397–405.(2). Uemura N., Okamoto S., Yamamoto S., Matsumura N., Yamaguchi S., Yamakido M., Taniyama K., Sasaki N., Schlemper R. J. N. Engl. J.Med. 2001; 345:784–789.(3). Boquet P., Ricci V., Galmiche A., Gauthier N. C.Trends Microbiol. 2003; 11:410–413.(4). Warren J. R., Marshall B.Lancet. 1983; 321:1273–1275.(5). Blaser M. J.EMBO Rep. 2006; 7:956–960.
Article(6). Dore M. P., Lu H., Graham D. Y.Gut. 2016; Epub ahead of print.(7). Wang J., Li W. T., Zheng Y. X., Zhao S. S., Li N., Huang Y., Zhou R. R., Huang Z. B., Fan X. G.Gastroenterol. Res. Pract. 2016; Epub ahead of print.(8). Malfertheiner P., Selgrad M.Curr. Opin. Gastroenterol. 2014; 30:589–595.(9). Dos Santos A. A., Carvalho A. A.World J. Gastroenterol. 2015; 21:139–154.(10). Austin A., Jegadeesan M., Gowrishankar R.Nat. Prod. Sci. 2003; 9:1–3.(11). Arichi H., Kimura Y., Okuda H., Baba K., Kozawa M., Arichi S.Chem. Pharm. Bull. 1980; 30:1766–1770.(12). Su P. W., Yang C. -H., Yang J. -F., Su P. Y., Chuang L. Y.Molecules. 2015; 20:11119–11130.(13). Zhang W. T., Jia Y., Huang Q. W., Li Q., Bi K. S.Chromatographia. 2007; 66:685–689.(14). Jiangsu New Medical College. Dictionary of Chinese Materia Medica; Science and Technology Press: China. 1977; 1329–1331.(15). Zhang H., Li C., Kwok S. T., Zhang Q. W., Chan S. W.Evid. Based Complement. Alternat. Med. 2013; 208349.(16). Peng W., Qin R., Li X., Zhou H. J.Ethnopharmacol. 2013; 148:729–745.(17). Lin C. J., Lin H. J., Chen T. H., Hsu Y. A., Liu C. S., Hwang G. Y., Wan L.PLoS One. 2015; 10:e0117602.(18). Lee C. C., Chen Y. T., Chiu C. C., Liao W. T., Liu Y. C., David Wang H. M. J.Biosci. Bioeng. 2015; 119:464–469.(19). Su P. W., Yang C. -H., Yang J. -F., Su P. Y., Chuang L. Y.Molecules. 2015; 20:11119–11130.(20). Park W. S., Bae J. -Y., Kim H. J., Kim M. K., Lee W. K., Kang H. -L., Baik S. C., Lim K. M., Lee M. K., Ahn M.-J. Nat. Prod. Sci. 2015; 21:49–53.(21). Amin M., Anwer M., Naz F., Mehmood T., Saari N.Molecules. 2013; 18:2135–2149.(22). Yang Lu. In Introduction to Natural Product Chemistry: Anthraquinones. Renheng X. U., Yang Y., Zhao W., editorsCRC press;USA: 2012; 10:189–203.(23). Chu X., Sun A., Liu R. J.Chromatogr. A. 2005; 1097:33–39.(24). Sivakumar B., Murugan R., Baskaran A., Khadangale B. P., Murugan S., Senthilkumar U. P.Sci. Pharm. 2013; 81:683–695.(25). Zhang W., Ye M., Zhan J., Chen Y., Guo D.Biotechnol. Lett. 2004; 26:127–131.(26). Kimura Y., Kozawa M., Baba K., Hata K.Planta Med. 1983; 48:164–168.(27). Brown J. C., Wang J., Kasman L., Jiang X., Haley-Zitlin V. J.Appl. Microbiol. 2011; 110:139–146.(28). Park B. S., Lee H. K., Lee S. E., Piao X. L., Takeoka G. R., Wong R. Y., Ahn Y. J., Kim J. H. J.Ethnopharmacol. 2006; 105:255–262.(29). Skouloubris S., Djaout K., Lamarre I., Lambry J. C., Anger K., Briffotaux J., Liebl U., de Reuse, H. and Myllykallio H.Open Biol. 2015; 5:150015.(30). Lee I. -S., Im H. G., Lee S.Korean J. Food Sci. Technol. 2003; 35:1182–1187.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Study on the antimicrobial activities of herbal extracts against Helicobacter pylori
- Anti-Helicobacter pylori Activity of Compounds Isolated from Fraxinus mandshurica Bark
- Future of Helicobacter pylori Eradication-New Trials
- Virtual Screening of a Series of Phytocompounds from Polygonum cuspidatum for Identification of Potential Antibacterial Drug Candidates: an In-silico and Drug Design Approaches
- Response to Treatment of Helicobacter pylori-associated Dyspepsia: Eradication of Helicobacter pylori or Correction of Gastric or Intestinal Dysbiosis?