Restor Dent Endod.
2015 Feb;40(1):14-22. 10.5395/rde.2015.40.1.14.
Epigenetics: general characteristics and implications for oral health
- Affiliations
-
- 1Department of Conservative Dentistry, Seoul National University School of Dentistry and Dental Research Institute, Seoul, Korea. dgseo@snu.ac.kr
- 2Department of Nutritional Science and Food Management, Ewha Womans University, Seoul, Korea.
- 3Department of Dentistry, Inje University Seoul Paik Hospital, Seoul, Korea.
- 4Nutrition Education Major, Graduate School of Education, Sangmyung University, Seoul, Korea.
- KMID: 2316919
- DOI: http://doi.org/10.5395/rde.2015.40.1.14
Abstract
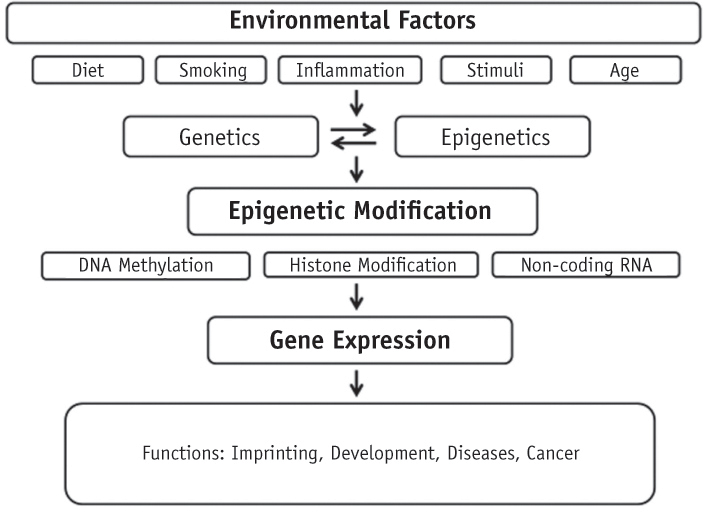
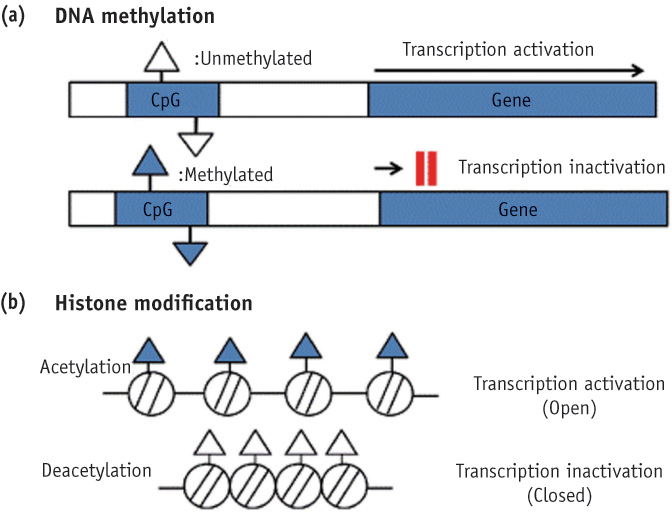
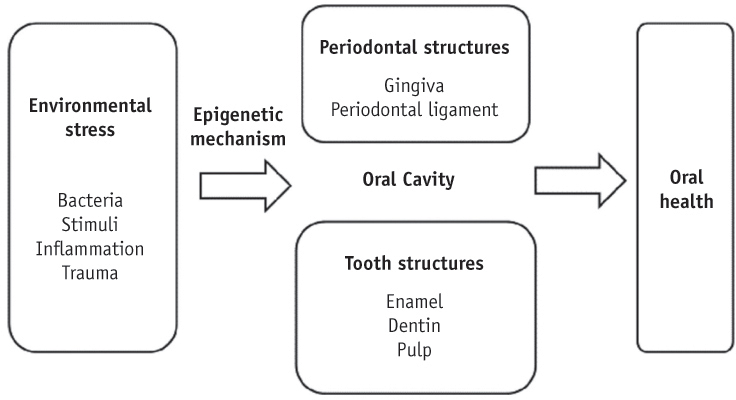
- Genetic information such as DNA sequences has been limited to fully explain mechanisms of gene regulation and disease process. Epigenetic mechanisms, which include DNA methylation, histone modification and non-coding RNAs, can regulate gene expression and affect progression of disease. Although studies focused on epigenetics are being actively investigated in the field of medicine and biology, epigenetics in dental research is at the early stages. However, studies on epigenetics in dentistry deserve attention because epigenetic mechanisms play important roles in gene expression during tooth development and may affect oral diseases. In addition, understanding of epigenetic alteration is important for developing new therapeutic methods. This review article aims to outline the general features of epigenetic mechanisms and describe its future implications in the field of dentistry.
MeSH Terms
Figure
Reference
-
1. Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007; 128:635–638.
Article2. Holliday R. Epigenetics: a historical overview. Epigenetics. 2006; 1:76–80.
Article3. Holliday R. Mechanisms for the control of gene activity during development. Biol Rev Camb Philos Soc. 1990; 65:431–471.
Article4. Russo VE, Martienssen RA, Riggs AD. Epigenetic mechanisms of gene regulation. New York: Cold Spring Harbor Laboratory Press;1996. p. 1–4.5. Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009; 23:781–783.6. Bayarsaihan D. Epigenetic mechanisms in inflammation. J Dent Res. 2011; 90:9–17.
Article7. Barros SP, Offenbacher S. Epigenetics: connecting environment and genotype to phenotype and disease. J Dent Res. 2009; 88:400–408.
Article8. Kaikkonen MU, Lam MT, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res. 2011; 90:430–440.
Article9. Lod S, Johansson T, Abrahamsson KH, Larsson L. The influence of epigenetics in relation to oral health. Int J Dent Hyg. 2014; 12:48–54.
Article10. Katsnelson A. Epigenome effort makes its mark. Nature. 2010; 467:646.
Article11. Williams SD, Hughes TE, Adler CJ, Brook AH, Townsend GC. Epigenetics: a new frontier in dentistry. Aust Dent J. 2014; 59:Suppl 1. 23–33.
Article12. Fan Z, Yamaza T, Lee JS, Yu J, Wang S, Fan G, Shi S, Wang CY. BCOR regulates mesenchymal stem cell function by epigenetic mechanisms. Nat Cell Biol. 2009; 11:1002–1009.
Article13. Brook AH. Multilevel complex interactions between genetic, epigenetic and environmental factors in the aetiology of anomalies of dental development. Arch Oral Biol. 2009; 54:Supplement 1. S3–S17.
Article14. Townsend GC, Richards L, Hughes T, Pinkerton S, Schwerdt W. Epigenetic influences may explain dental differences in monozygotic twin pairs. Aust Dent J. 2005; 50:95–100.
Article15. Townsend G, Bockmann M, Hughes T, Brook A. Genetic, environmental and epigenetic influences on variation in human tooth number, size and shape. Odontology. 2012; 100:1–9.
Article16. Gomez RS, Dutra WO, Moreira PR. Epigenetics and periodontal disease: future perspectives. Inflamm Res. 2009; 58:625–629.
Article17. Lindroth AM, Park YJ. Epigenetic biomarkers: a step forward for understanding periodontitis. J Periodontal Implant Sci. 2013; 43:111–120.
Article18. Zhang S, Crivello A, Offenbacher S, Moretti A, Paquette DW, Barros SP. Interferon-gamma promoter hypomethylation and increased expression in chronic periodontitis. J Clin Periodontol. 2010; 37:953–961.
Article19. Zhang S, Barros SP, Moretti AJ, Yu N, Zhou J, Preisser JS, Niculescu MD, Offenbacher S. Epigenetic regulation of TNFA expression in periodontal disease. J Periodontol. 2013; 84:1606–1616.20. Zhang S, Barros SP, Niculescu MD, Moretti AJ, Preisser JS, Offenbacher S. Alteration of PTGS2 promoter methylation in chronic periodontitis. J Dent Res. 2010; 89:133–137.
Article21. Kinane DF, Hart TC. Genes and gene polymorphisms associated with periodontal disease. Crit Rev Oral Biol Med. 2003; 14:430–449.
Article22. Stenvinkel P, Karimi M, Johansson S, Axelsson J, Suliman M, Lindholm B, Heimbürger O, Barany P, Alvestrand A, Nordfors L, Qureshi AR, Ekström TJ, Schalling M. Impact of inflammation on epigenetic DNA methylation - a novel risk factor for cardiovascular disease? J Intern Med. 2007; 261:488–499.
Article23. Cardoso FP, Viana MB, Sobrinho AP, Diniz MG, Brito JA, Gomes CC, Moreira PR, Gomez RS. Methylation pattern of the IFN-gamma gene in human dental pulp. J Endod. 2010; 36:642–646.24. Cardoso FP, de Faria Amormino SA, Dutra WO, Ribeiro Sobrinho AP, Moreira PR. Methylation pattern of the CD14 and TLR2 genes in human dental pulp. J Endod. 2014; 40:384–386.25. Duncan HF, Smith AJ, Fleming GJ, Cooper PR. Histone deacetylase inhibitors induced differentiation and accelerated mineralization of pulp-derived cells. J Endod. 2012; 38:339–345.
Article26. Duncan HF, Smith AJ, Fleming GJ, Cooper PR. Histone deacetylase inhibitors epigenetically promote reparative events in primary dental pulp cells. Exp Cell Res. 2013; 319:1534–1543.
Article27. Hui T, A P, Zhao Y, Wang C, Gao B, Zhang P, Wang J, Zhou X, Ye L. EZH2, a potential regulator of dental pulp inflammation and regeneration. J Endod. 2014; 40:1132–1138.
Article28. Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009; 10:805–811.
Article29. Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000; 405:486–489.30. Loenen WA. S-adenosylmethionine: jack of all trades and master of everything? Biochem Soc Trans. 2006; 34:330–333.
Article31. Vucic EA, Brown CJ, Lam WL. Epigenetics of cancer progression. Pharmacogenomics. 2008; 9:215–234.
Article32. Bird AP, Wolffe AP. Methylation-induced repression-belts, braces, and chromatin. Cell. 1999; 99:451–454.
Article33. Cheung HH, Lee TL, Rennert OM, Chan WY. DNA methylation of cancer genome. Birth Defects Res C Embryo Today. 2009; 87:335–350.
Article34. Fuchs J, Demidov D, Houben A, Schubert I. Chromosomal histone modification patterns-from conservation to diversity. Trends Plant Sci. 2006; 11:199–208.
Article35. Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009; 43:559–599.
Article36. Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007; 129:823–837.
Article37. Lan F, Shi Y. Epigenetic regulation: methylation of histone and non-histone proteins. Sci China C Life Sci. 2009; 52:311–322.
Article38. Duncan HF, Smith AJ, Fleming GJ, Cooper PR. HDACi: cellular effects, opportunities for restorative dentistry. J Dent Res. 2011; 90:1377–1388.39. Paino F, La Noce M, Tirino V, Naddeo P, Desiderio V, Pirozzi G, De Rosa A, Laino L, Altucci L, Papaccio G. Histone deacetylase inhibition with valproic acid downregulates osteocalcin gene expression in human dental pulp stem cells and osteoblasts: evidence for HDAC2 involvement. Stem Cells. 2014; 32:279–289.
Article40. Wang T, Liu H, Ning Y, Xu Q. The histone acetyltransferase p300 regulates the expression of pluripotency factors and odontogenic differentiation of human dental pulp cells. PLoS One. 2014; 9:e102117.
Article41. Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010; 466:835–840.
Article42. Sun Q, Liu H, Chen Z. The fine tuning role of microRNA-RNA interaction in odontoblast differentiation and disease. Oral Dis. 2014; 03. 22. doi: 10.1111/odi.12237. [Epub ahead of print].
Article43. Perez P, Jang SI, Alevizos I. Emerging landscape of non-coding RNAs in oral health and disease. Oral Dis. 2014; 20:226–235.
Article44. Reik W, Walter J. Imprinting mechanisms in mammals. Curr Opin Genet Dev. 1998; 8:154–164.
Article45. Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999; 99:247–257.
Article46. Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007; 213:384–390.
Article47. Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007; 447:425–432.
Article48. Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol. 2003; 4:616–623.
Article49. Post WS, Goldschmidt-Clermont PJ, Wilhide CC, Heldman AW, Sussman MS, Ouyang P, Milliken EE, Issa JP. Methylation of the estrogen receptor gene is associated with aging and atherosclerosis in the cardiovascular system. Cardiovasc Res. 1999; 43:985–991.
Article50. Lund G, Andersson L, Lauria M, Lindholm M, Fraga MF, Villar-Garea A, Ballestar E, Esteller M, Zaina S. DNA methylation polymorphisms precede any histological sign of atherosclerosis in mice lacking apolipoprotein E. J Biol Chem. 2004; 279:29147–29154.
Article51. Zaina S, Lindholm MW, Lund G. Nutrition and aberrant DNA methylation patterns in atherosclerosis: more than just hyperhomocysteinemia? J Nutr. 2005; 135:5–8.
Article52. Barker DJ, Eriksson JG, Forsén T, Osmond C. Fetal origins of adult disease: strength of effects and biological basis. Int J Epidemiol. 2002; 31:1235–1239.
Article53. Razin A, Shemer R. DNA methylation in early development. Hum Mol Genet. 1995; 4:1751–1755.
Article54. Ohi T, Uehara Y, Takatsu M, Watanabe M, Ono T. Hypermethylation of CpGs in the promoter of the COL1A1 gene in the aged periodontal ligament. J Dent Res. 2006; 85:245–250.
Article55. Wu H, Lippmann JE, Oza JP, Zeng M, Fives-Taylor P, Reich NO. Inactivation of DNA adenine methyltransferase alters virulence factors in Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 2006; 21:238–244.
Article56. Ito K. Impact of post-translational modifications of proteins on the inflammatory process. Biochem Soc Trans. 2007; 35:281–283.
Article57. Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003; 349:2042–2054.
Article58. Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004; 4:143–153.
Article59. Breivik J, Gaudernack G. Genomic instability, DNA methylation, and natural selection in colorectal carcinogenesis. Semin Cancer Biol. 1999; 9:245–254.
Article60. Choi S, Myers JN. Molecular pathogenesis of oral squamous cell carcinoma: implications for therapy. J Dent Res. 2008; 87:14–32.
Article61. Babel N, Cherepnev G, Babel D, Tropmann A, Hammer M, Volk HD, Reinke P. Analysis of tumor necrosis factor-alpha, transforming growth factor-beta, interleukin-10, IL-6, and interferon-gamma gene polymorphisms in patients with chronic periodontitis. J Periodontol. 2006; 77:1978–1983.
Article62. Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006; 5:769–784.
Article63. Balasubramanian S, Verner E, Buggy JJ. Isoform-specific histone deacetylase inhibitors: the next step? Cancer Lett. 2009; 280:211–221.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Inflammaging: Molecular Pathways and Implications in Oral Pathology
- Effects of oral health-related self-efficacy on oral health-related quality of life in male high school students
- Experience, educational needs and perception of dentine hypersensitivity of dental hygienists
- Asthma and epigenetics
- A study on international oral health policy: implications from the 13 th Asian Chief Dental Officers’ Meeting