Ann Dermatol.
2010 Feb;22(1):41-47. 10.5021/ad.2010.22.1.41.
Molecular Analysis of Malassezia Microflora on the Skin of the Patients with Atopic Dermatitis
- Affiliations
-
- 1Department of Dermatology, Konkuk University School of Medicine, Seoul, Korea. kjahn@kuh.ac.kr
- 2Department of Dermatology, Konkuk University Research Institute of Medical Science, Seoul, Korea.
- KMID: 2172035
- DOI: http://doi.org/10.5021/ad.2010.22.1.41
Abstract
- BACKGROUND
The yeasts of the genus Malassezia are members of the normal flora on human skin and they are found in 75~80% of healthy adults. Since its association with various skin disorders have been known, there have been a growing number of reports that have implicated Malassezia yeast in atopic dermatitis (AD).
OBJECTIVE
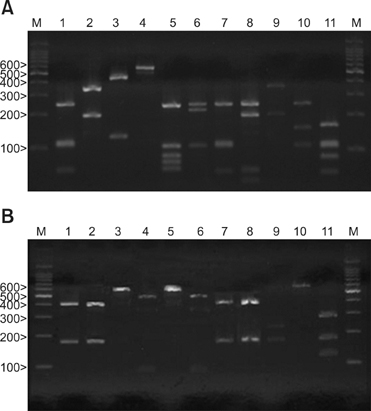
The aim of the present study is to isolate the various Malassezia species from AD patients by using 26S rDNA (ribosomal Deoxyribonucleic acid) PCR-RFLP and to investigate the relationship between a positive Malassezia culture and the severity of AD.
METHODS
Cultures for Malassezia yeasts were taken from the scalp, cheek, chest, arm and thigh of 60 patients with atopic dermatitis. We used a rapid and accurate molecular biological method 26S rDNA PCR-RFLP, and this method can overcome the limits of the morphological and biochemical methods. RESULTS: Positive Malassezia growth was noted on 51.7% of the patients with atopic dermatitis by 26S rDNA PCR-RFLP analysis. The overall dominant species was M. sympodialis (16.3%). M. restricta was the most common species on the scalp (30.0%) and cheek (16.7%). M. sympodialis (28.3%) was the most common species on the chest. The positive culture rate was the highest for the 11~20 age group (59.0%) and the scalp showed the highest rate at 66.7%. There was no significant relationship between the Malassezia species and SCORing for Atopic Dermatitis (SCORAD).
CONCLUSION
The fact that the cultured species was different for the atopic dermatitis lesion skin from that of the normal skin may be due to the disrupted skin barrier function and sensitization of the organism induced by scratching in the AD lesion-skin. But there was no relationship between the Malassezia type and the severity score. The severity score is thought to depend not on the type, but also on the quantity of the yeast.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Progress in Malassezia Research in Korea
Soo Young Kim, Yang Won Lee, Yong Beom Choe, Kyu Joong Ahn
Ann Dermatol. 2015;27(6):647-657. doi: 10.5021/ad.2015.27.6.647.
Reference
-
1. Ahn KJ. Taxonomy of the genus Malassezia. Korean J Med Mycol. 1998. 3:81–88.2. Ljubojevic S, Skerlev M, Lipozencic J, Basta-Juzbasic A. The role of Malassezia furfur in dermatology. Clin Dermatol. 2002. 20:179–182.3. Baker BS. The role of microorganisms in atopic dermatitis. Clin Exp Immunol. 2006. 144:1–9.
Article4. Nikkels AF, Pierard GE. Framing the future of antifungals in atopic dermatitis. Dermatology. 2003. 206:398–400.
Article5. Faergemann J. Atopic dermatitis and fungi. Clin Microbiol Rev. 2002. 15:545–563.
Article6. Aspres N, Anderson C. Malassezia yeasts in the pathogenesis of atopic dermatitis. Australas J Dermatol. 2004. 45:199–207.
Article7. Hanifin JM, Rajka G. Diagnostic features of atopic dermatitis. Acta Derm Venereol. 1980. 92:Suppl. 44–47.8. Leeming JP, Notman FH, Holland KT. The distribution and ecology of Malassezia furfur and cutaneous bacteria on human skin. J Appl Bacteriol. 1989. 67:47–52.
Article9. Leeming JP, Notman FH. Improved methods for isolation and enumeration of Malassezia furfur from human skin. J Clin Microbiol. 1987. 25:2017–2019.
Article10. Mirhendi H, Makimura K, Zomorodian K, Yamada T, Sugita T, Yamaguchi H. A simple PCR-RFLP method for identification and differentiation of 11 Malassezia species. J Microbiol Methods. 2005. 61:281–284.
Article11. Crespo Erchiga V, Ojeda Martos A, Vera Casano A, Crespo Erchiga A, Sanchez Fajardo F. Malassezia globosa as the causative agent of pityriasis versicolor. Br J Dermatol. 2000. 143:799–803.12. Gupta AK, Kohli Y, Faergemann J, Summerbell RC. Epidemiology of Malassezia yeasts associated with pityriasis ersicolor in Ontario, Canada. Med Mycol. 2001. 39:199–206.
Article13. Gupta AK, Kohli Y, Summerbell RC, Faergemann J. Quantitative culture of Malassezia species from different body sites of individuals with or without dermatoses. Med Mycol. 2001. 39:243–251.
Article14. Kim SC, Kim HU. The distribution of Malassezia species on the normal human skin according to body region. Korean J Med Mycol. 2000. 5:120–128.15. Nakabayashi A, Sei Y, Guillot J. Identification of Malassezia species isolated from patients with seborrhoeic dermatitis, atopic dermatitis, pityriasis versicolor and normal subjects. Med Mycol. 2000. 38:337–341.
Article16. Sugita T, Suto H, Unno T, Tsuboi R, Ogawa H, Shinoda T, et al. Molecular analysis of Malassezia microflora on the skin of atopic dermatitis patients and healthy subjects. J Clin Microbiol. 2001. 39:3486–3490.
Article17. Lee YW, Yim SM, Lim SH, Choe YB, Ahn KJ. Quantitative investigation on the distribution of Malassezia species on healthy human skin in Korea. Mycoses. 2006. 49:405–410.
Article18. Jang SJ, Lim SH, Ko JH, Oh BH, Kim SM, Song YC, et al. The investigation on the distribution of Malassezia yeasts on the normal Korean skin by 26S rDNA PCR-RFLP. Ann Dermatol. 2009. 21:18–26.
Article19. Schafer L, Kragballe K. Abnormalities in epidermal lipid metabolism in patients with atopic dermatitis. J Invest Dermatol. 1991. 96:10–15.
Article20. Sandstrom Falk MH, Tengvall Linder M, Johansson C, Bartosik J, Back O, Sarnhult T, et al. The prevalence of Malassezia yeasts in patients with atopic dermatitis, seborrhoeic dermatitis and healthy controls. Acta Derm Venereol. 2005. 85:17–23.21. Sugita T, Takashima M, Shinoda T, Suto H, Unno T, Tsuboi R, et al. New yeast species, Malassezia dermatis, isolated from patients with atopic dermatitis. J Clin Microbiol. 2002. 40:1363–1367.
Article22. Scheynius A, Johansson C, Buentke E, Zargari A, Linder MT. Atopic eczema/dermatitis syndrome and Malassezia. Int Arch Allergy Immunol. 2002. 127:161–169.23. Takahata Y, Sugita T, Kato H, Nishikawa A, Hiruma M, Muto M. Cutaneous Malassezia flora in atopic dermatitis differs between adults and children. Br J Dermatol. 2007. 157:1178–1182.
Article24. Lee YW, Lim SH, Ahn KJ. The application of 26S rDNA PCR-RFLP in the identification and classification of Malassezia yeast. Korean J Med Mycol. 2006. 11:141–153.25. Oh BH, Song YC, Lee YW, Choe YB, Ahn KJ. Comparison of nested PCR and RFLP for identification and classification of Malassezia yeasts from healthy human skin. Ann Dermatol. 2009. 21:352–357.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Progress in Malassezia Research in Korea
- Comparison of Clinical Manifestations and Distribution of Malassezia Species in Patients with Seborrheic Dermatitis and Atopic Dermatitis
- Difference of Malassezia Species and Pityrosporum ovale Specific IgE in Head and Neck Lesions of Atopic Dermatitis Related to Ages and Severity
- Comparison of Nested PCR and RFLP for Identification and Classification of Malassezia Yeasts from Healthy Human Skin
- In vitro Susceptibility of the Six Malassezia Species to Itraconazole, Fluconazole and Terbinafine