Anat Cell Biol.
2010 Mar;43(1):78-85. 10.5115/acb.2010.43.1.78.
Chromatin organization and transcriptional activation of Hox genes
- Affiliations
-
- 1Department of Anatomy, Embryology Laboratory, Yonsei University College of Medicine, Seoul, Korea. mhkim1@yuhs.ac
- 2Brain Korea 21 Project for Biomedical Science, Yonsei University College of Medicine, Seoul, Korea.
- KMID: 2168900
- DOI: http://doi.org/10.5115/acb.2010.43.1.78
Abstract
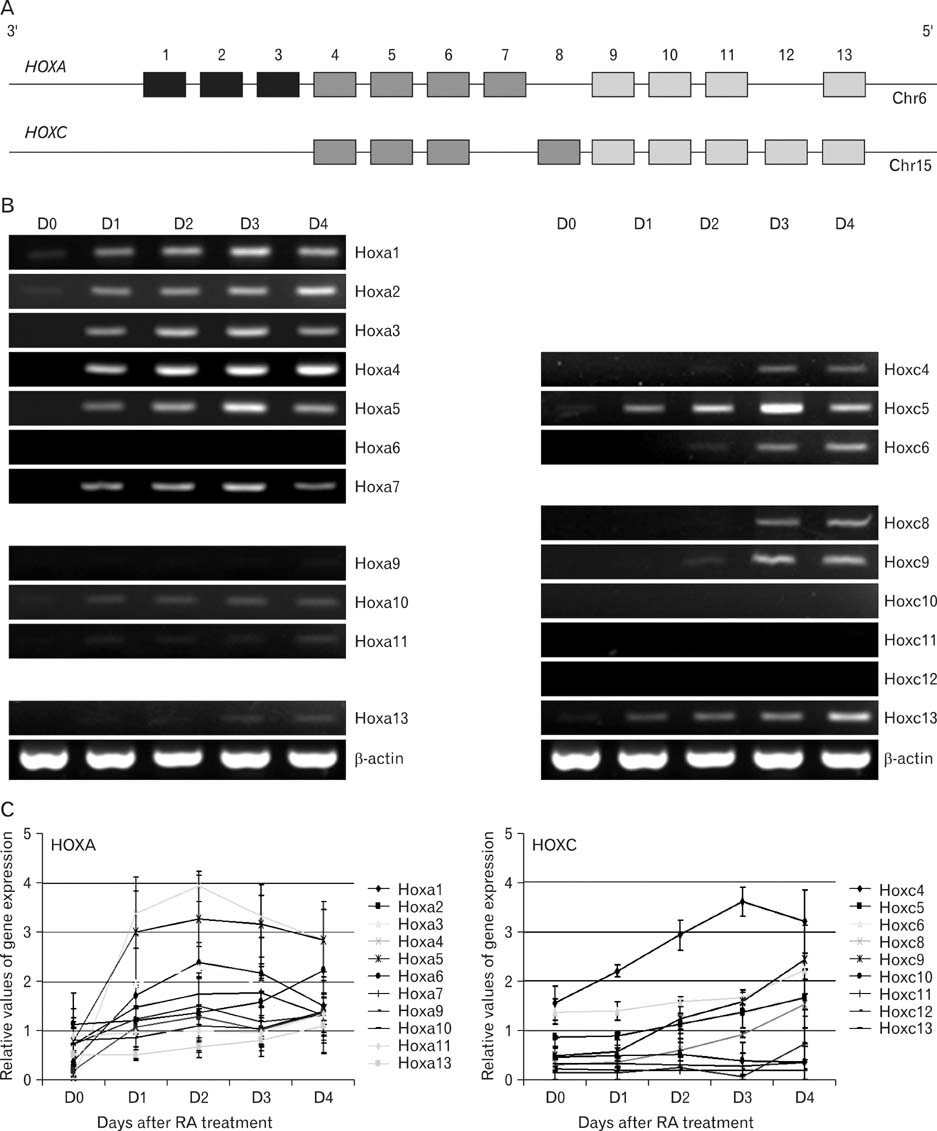
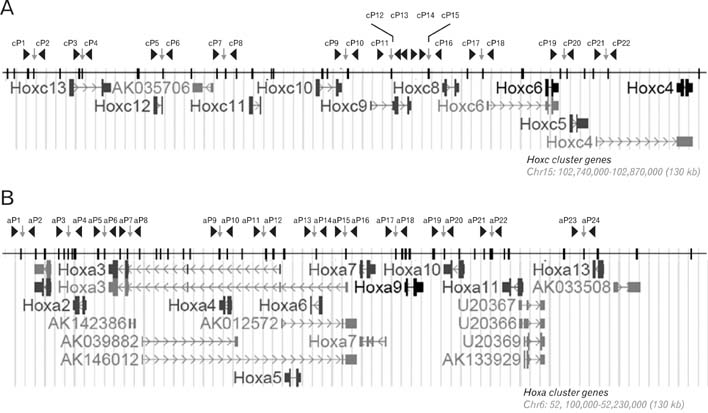
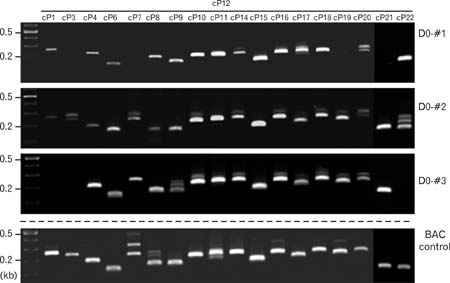
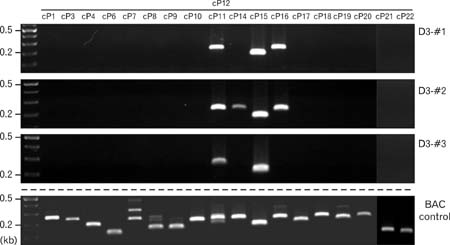
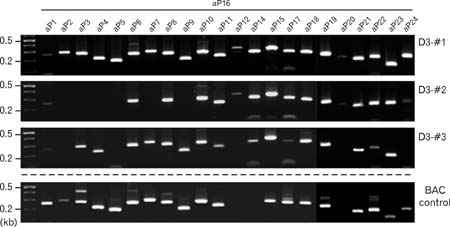
- Spatially and temporally programmed expression of the Hox genes along the antero-posterior (A-P) axis is essential for correct pattern formation during embryonic development. An accumulating body of evidence indicates the pivotal role of spatial chromatin organization for the coordination of gene regulation. Recently, chromosome conformation capture (3C) technique has been developed and opened a new way to study chromosomal interactions in the nucleus. In this study, we describe 3C method we applied in F9 embryonic teratocarcinoma cells and demonstrate that the chromosomal interactions at Hox loci are successfully detected. Interestingly, at Hoxc loci, the abundance of intrachromosomal interactions with neighboring fragments was drastically decreased when the genes are expressed. These results indicate the possibility of the dynamic pattern of chromosomal interaction in association with the transcriptional regulation of Hox genes.
Keyword
MeSH Terms
Figure
Reference
-
1. Boncinelli E, Simeone A, Acampora D, Mavilio F. HOX gene activation by retinoic acid. Trends Genet. 1991. 7:329–334.2. Boney-Montoya J, Ziegler YS, Curtis CD, Montoya JA, Nardulli AM. Long-Range transcriptional control of progesterone receptor gene expression. Mol Endocrinol. 2010. 24:346–358.3. Breier G, Bućan M, Francke U, Colberg-Poley AM, Gruss P. Sequential expression of murine homeo box genes during F9 EC cell differentiation. EMBO J. 1986. 5:2209–2215.4. Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002. 32:623–626.5. Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004. 18:1119–1130.6. Chavanas S, Adoue V, Mechin MC, et al. Long-range enhancer associated with chromatin looping allows AP-1 regulation of the peptidylarginine deiminase 3 gene in differentiated keratinocyte. PLoS ONE. 2008. 3:e3408.7. Dekker J. Ggene regulation in the third dimension. Science. 2008. 319:1793–1794.8. Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002. 295:1306–1311.9. Fraser J, Rousseau M, Shenker S, et al. Chromatin conformation signatures of cellular differentiation. Genome Biology. 2009. 10:R37.10. Gaunt SJ, Strachan L. Temporal colinearity in expression of anterior hox genes in developing chick embryos. Dev Dyn. 1996. 207:270–280.11. Izpisúa-Belmonte JC, Falkenstein H, Dollé P, Renucci A, Duboule D. Murine genes related to the Drosophila AbdB homeotic genes are sequentially expressed during development of the posterior part of the body. EMBO J. 1991. 10:2279–2289.12. Kleinjan DA, Lettice LA, Veronica van H, Robert EH. Chapter 13 long range gene control and genetic disease. Advances in genetics, academic press. 2008. Volume 61:339–388.13. Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006. 126:403–413.14. Murphy SP, Garbern J, Odenwald WF, Lazzarini RA, Linney E. Differential expression of the homeobox gene Hox-1.3 in F9 embryonal carcinoma cells. Proc Natl Acad Sci USA. 1988. 85:5587–5591.15. Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004. 36:889–893.16. Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol. 2004. 5:1017–1027.17. Stornaiuolo A, Acampora D, Pannese M, et al. Human HOX genes are differentially activated by retinoic acid in embryonal carcinoma cells according to their position within the four loci. Cell Differ Dev. 1990. 31:119–127.18. Tolhuis B, Palstra R-J, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002. 10:1453–1465.19. Wang KC, Helms JA, Chang HY. Regeneration, repair and remembering identity: the three Rs of Hox gene expression. Trends Cell Biol. 2009. 19:268–275.20. Würtele H, Chartrand P. Genome-wide scanning of HoxB1-associated loci in mouse ES cells using an open-ended chromosome conformation capture methodology. Chromosome Res. 2006. 14:477–495.21. Yu SJ, Lee JY, Kim SH, Deocaris CC, Kim MH. Synthetic maternal stress hormone can modulate the expression of Hox genes. J Exp Biomed Sci. 2009. 15:249–255.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Influence of Polycomb Proteins and Epigenetic Transcriptional Modifiers on the Development and Activation of T Lymphocytes
- The Function of the Vitamin D Receptor and a Possible Role of Enhancer RNA in Epigenomic Regulation of Target Genes: Implications for Bone Metabolism
- Epigenetics and Psychiatric Disorders
- Effects of estrogen receptor and estrogen on the chromatin structure in estrogen receptor stable transfectants
- Epigenetic regulation and chromatin remodeling in learning and memory