Ann Surg Treat Res.
2015 Aug;89(2):74-80. 10.4174/astr.2015.89.2.74.
Association between genetic polymorphisms in cortactin and susceptibility to gastric cancer
- Affiliations
-
- 1Department of Surgery, Institute of Medical Science, Wonkwang University School of Medicine, Iksan, Korea. rjk@wonkwang.ac.kr
- 2Department of Pathology, Institute of Medical Science, Wonkwang University School of Medicine, Iksan, Korea.
- KMID: 2166974
- DOI: http://doi.org/10.4174/astr.2015.89.2.74
Abstract
- PURPOSE
Overexpression of cortactin (CTTN) in human tumors has been proposed to result in increased cell migration and metastatic potential. Here, we determined the frequencies of CTTN g.-9101C>T, g.-8748C>T, and g.72C>T polymorphisms in apparently healthy subjects and gastric cancer patients, respectively, and the influence of the CTTN polymorphisms on gastric cancer susceptibility.
METHODS
Blood samples were collected from 267 patients and 533 controls. CTTN g.-8748C>T and g.-9101C>T polymorphisms were determined using polymerase chain reaction-restriction fragment length polymorphism; the g.72C>T polymorphism was determined using the TaqMan method.
RESULTS
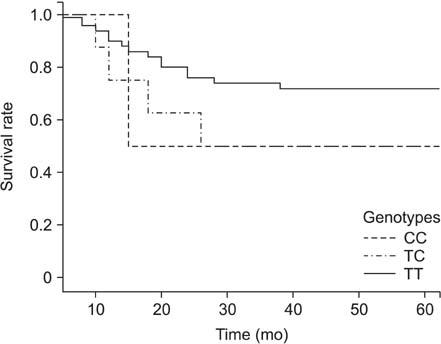
Genotype frequencies of the CTTN g.-9101C>T polymorphism were 97.5% (TT), 2.5% (TC), and 0% (CC) in the patient group, and 98.6% (TT), 1.4% (TC), and 0% (CC) in the control group. Genotype frequencies of the CTTN g.-8748C>T polymorphism were 93.3% (TT), 6.8% (TC), and 0% (CC) in the patient group, and 94.2% (TT), 5.8% (TC), and 0% (CC) in the control group. Genotype frequencies of the CTTN g.72C>T polymorphism were 82.4% (CC), 17.2% (CT), and 0.4% (TT) in the patient group, and 78.0% (CC), 20.1% (CT), and 1.9% (TT) in the control group. Genotype and allele frequencies of the CTTN g.-9101C>T polymorphism differed significantly between the advanced gastric cancer and control groups. Patients with advanced gastric cancer, possessing the TC genotype, had a significantly poorer prognosis than the group with the TT genotype.
CONCLUSION
The CTTN g.-9101C>T polymorphism might influence advanced gastric cancer susceptibility. However, the role of the CTTN g.-9101C>T, g.-8748C>T, and g.72C>T polymorphisms requires careful interpretation and confirmation through larger studies.
MeSH Terms
Figure
Reference
-
1. The Information Committee of the Korean Gastric Cancer Association. 2004 Nationwide Gastric Cancer Report in Korea. J Korean Gastric Cancer Assoc. 2007; 7:47–54.2. Hartgrink HH, Jansen EP, van Grieken NC, van de Velde CJ. Gastric cancer. Lancet. 2009; 374:477–490.3. de Martel C, Forman D, Plummer M. Gastric cancer: epidemiology and risk factors. Gastroenterol Clin North Am. 2013; 42:219–240.4. Wang XQ, Yan H, Terry PD, Wang JS, Cheng L, Wu WA, et al. Interaction between dietary factors and Helicobacter pylori infection in noncardia gastric cancer: a population-based case-control study in China. J Am Coll Nutr. 2012; 31:375–384.5. Songun I, van de Velde CJ, Arends JW, Blok P, Grond AJ, Offerhaus GJ, et al. Classification of gastric carcinoma using the Goseki system provides prognostic information additional to TNM staging. Cancer. 1999; 85:2114–2118.6. Shiraishi N, Sato K, Yasuda K, Inomata M, Kitano S. Multivariate prognostic study on large gastric cancer. J Surg Oncol. 2007; 96:14–18.7. Bougneres L, Girardin SE, Weed SA, Karginov AV, Olivo-Marin JC, Parsons JT, et al. Cortactin and Crk cooperate to trigger actin polymerization during Shigella invasion of epithelial cells. J Cell Biol. 2004; 166:225–235.8. Daly RJ. Cortactin signalling and dynamic actin networks. Biochem J. 2004; 382(Pt 1):13–25.9. van Rossum AG, Moolenaar WH, Schuuring E. Cortactin affects cell migration by regulating intercellular adhesion and cell spreading. Exp Cell Res. 2006; 312:1658–1670.10. van Rossum AG, Gibcus J, van der Wal J, Schuuring E. Cortactin overexpression results in sustained epidermal growth factor receptor signaling by preventing ligand-induced receptor degradation in human carcinoma cells. Breast Cancer Res. 2005; 7:235–237.11. Luo ML, Shen XM, Zhang Y, Wei F, Xu X, Cai Y, et al. Amplification and overexpression of CTTN (EMS1) contribute to the metastasis of esophageal squamous cell carcinoma by promoting cell migration and anoikis resistance. Cancer Res. 2006; 66:11690–11699.12. Rothschild BL, Shim AH, Ammer AG, Kelley LC, Irby KB, Head JA, et al. Cortactin overexpression regulates actinrelated protein 2/3 complex activity, motility, and invasion in carcinomas with chromosome 11q13 amplification. Cancer Res. 2006; 66:8017–8025.13. Akervall JA, Jin Y, Wennerberg JP, Zatterstrom UK, Kjellen E, Mertens F, et al. Chromosomal abnormalities involving 11q13 are associated with poor prognosis in patients with squamous cell carcinoma of the head and neck. Cancer. 1995; 76:853–859.14. Chuma M, Sakamoto M, Yasuda J, Fujii G, Nakanishi K, Tsuchiya A, et al. Overexpression of cortactin is involved in motility and metastasis of hepatocellular carcinoma. J Hepatol. 2004; 41:629–636.15. Tsai WC, Jin JS, Chang WK, Chan DC, Yeh MK, Cherng SC, et al. Association of cortactin and fascin-1 expression in gastric adenocarcinoma: correlation with clinicopathological parameters. J Histochem Cytochem. 2007; 55:955–962.16. Lee YY, Yu CP, Lin CK, Nieh S, Hsu KF, Chiang H, et al. Expression of survivin and cortactin in colorectal adenocarcinoma: association with clinicopathological parameters. Dis Markers. 2009; 26:9–18.17. Zhang K, Zhou B, Wang Y, Rao L, Zhang L. The TLR4 gene polymorphisms and susceptibility to cancer: a systematic review and meta-analysis. Eur J Cancer. 2013; 49:946–954.18. Saeki N, Ono H, Sakamoto H, Yoshida T. Genetic factors related to gastric cancer susceptibility identified using a genomewide association study. Cancer Sci. 2013; 104:1–8.19. Lynch HT, Grady W, Suriano G, Huntsman D. Gastric cancer: new genetic developments. J Surg Oncol. 2005; 90:114–133.20. Salisbury BA, Pungliya M, Choi JY, Jiang R, Sun XJ, Stephens JC. SNP and haplotype variation in the human genome. Mutat Res. 2003; 526:53–61.21. Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010; 17:1471–1474.22. Balasubramanian SP, Cox A, Brown NJ, Reed MW. Candidate gene polymorphisms in solid cancers. Eur J Surg Oncol. 2004; 30:593–601.23. Wu H, Parsons JT. Cortactin, an 80/85-kilodalton pp60src substrate, is a filamentous actin-binding protein enriched in the cell cortex. J Cell Biol. 1993; 120:1417–1426.24. Uruno T, Liu J, Zhang P, Fan Yx, Egile C, Li R, et al. Activation of Arp2/3 complexmediated actin polymerization by cortactin. Nat Cell Biol. 2001; 3:259–266.25. Timpson P, Lynch DK, Schramek D, Walker F, Daly RJ. Cortactin overexpression inhibits ligand-induced downregulation of the epidermal growth factor receptor. Cancer Res. 2005; 65:3273–3280.26. Clark ES, Whigham AS, Yarbrough WG, Weaver AM. Cortactin is an essential regulator of matrix metalloproteinase secretion and extracellular matrix degradation in invadopodia. Cancer Res. 2007; 67:4227–4235.27. Ormandy CJ, Musgrove EA, Hui R, Daly RJ, Sutherland RL. Cyclin D1, EMS1 and 11q13 amplification in breast cancer. Breast Cancer Res Treat. 2003; 78:323–335.28. Schuuring E. The involvement of the chromosome 11q13 region in human malignancies: cyclin D1 and EMS1 are two new candidate oncogenes: a review. Gene. 1995; 159:83–96.29. Zhao G, Huang ZM, Kong YL, Wen DQ, Li Y, Ren L, et al. Cortactin is a sensitive biomarker relative to the poor prognosis of human hepatocellular carcinoma. World J Surg Oncol. 2013; 11:74.30. Lee SY, Kang DB, Park WC, Lee JK, Chae SC. Association of CTTN polymorphisms with the risk of colorectal cancer. J Korean Surg Soc. 2012; 82:156–164.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Analysis of CYP1A1 , GST-mu and GST-theta Polymorphisms for the Determination of Genetic Susceptibility to Korean Gastric Cancer
- Gastric Cancer Susceptibility according to Methylenetetrahydrofolate Reductase and Thymidylate Synthase Gene Polymorphism
- Effects of Polymorphisms of Innate Immunity Genes and Environmental Factors on the Risk of Noncardia Gastric Cancer
- Association of RNase3 Polymorphisms with the Susceptibility of Gastric Cancer
- Association of the Interleukin-1beta and Interleukin-1 Receptor Antagonist Genetic Polymorphism and Korean Gastric Cancer