Clin Endosc.
2013 Nov;46(6):611-619.
Application of Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Imaging Mass Spectrometry (MALDI-TOF IMS) for Premalignant Gastrointestinal Lesions
- Affiliations
-
- 1Digestive Disease Center, CHA Bundang Medical Center, CHA University, Seongnam, Korea. hahmkb@cha.ac.kr
- 2Cancer Prevention Research Center, CHA University, Seoul, Korea.
- 3Lee Gil Ya Cancer and Diabetes Institute, Gachon University, Incheon, Korea.
Abstract
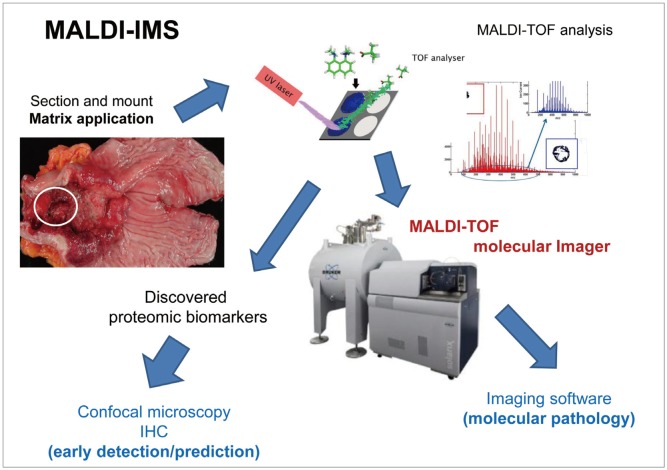
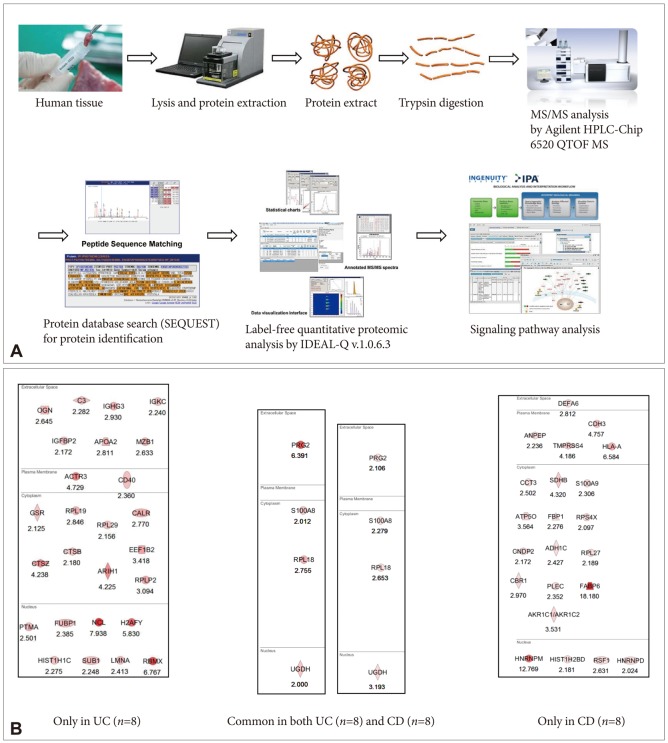
- Imaging mass spectrometry (IMS) is currently receiving large attention from the mass spectrometric community, although its use is not yet well known in the clinic. As matrix-assisted laser desorption/ionization time-of-flight (MALDI)-IMS can show the biomolecular changes in cells as well as tissues, it can be an ideal tool for biomedical diagnostics as well as the molecular diagnosis of clinical specimens, especially aimed at the prompt detection of premalignant lesions much earlier before overt mass formation, or for obtaining histologic clues from endoscopic biopsy. Besides its use for pathologic diagnosis, MALDI-IMS is also a powerful tool for the detection and localization of drugs, proteins, and lipids in tissue. Measurement of parameters that define and control the implications, challenges, and opportunities associated with the application of IMS to biomedical tissue studies might be feasible through a deep understanding of mass spectrometry. In this focused review series, new insights into the molecular processes relevant to IMS as well as other field applications are introduced.
Keyword
MeSH Terms
Figure
Reference
-
1. Yeo M, Kim DK, Park HJ, et al. Loss of transgelin in repeated bouts of ulcerative colitis-induced colon carcinogenesis. Proteomics. 2006; 6:1158–1165. PMID: 16402363.
Article2. Goetz M, Malek NP, Kiesslich R. Microscopic imaging in endoscopy: endomicroscopy and endocytoscopy. Nat Rev Gastroenterol Hepatol. 2013; 7. 30. Epub. DOI: 10.1038/nrgastro.2013.134.
Article3. Goetz M, Watson A, Kiesslich R. Confocal laser endomicroscopy in gastrointestinal diseases. J Biophotonics. 2011; 4:498–508. PMID: 21567975.
Article4. Alexandrov T. MALDI imaging mass spectrometry: statistical data analysis and current computational challenges. BMC Bioinformatics. 2012; 13(Suppl 16):S11. PMID: 23176142.
Article5. Ghafourian S, Sekawi Z, Raftari M, Ali MS. Application of proteomics in lab diagnosis. Clin Lab. 2013; 59:465–474. PMID: 23865343.
Article6. Tabb DL. Quality assessment for clinical proteomics. Clin Biochem. 2013; 46:411–420. PMID: 23246537.
Article7. Apweiler R, Aslanidis C, Deufel T, et al. Approaching clinical proteomics: current state and future fields of application in cellular proteomics. Cytometry A. 2009; 75:816–832. PMID: 19739086.
Article8. Azad NS, Rasool N, Annunziata CM, Minasian L, Whiteley G, Kohn EC. Proteomics in clinical trials and practice: present uses and future promise. Mol Cell Proteomics. 2006; 5:1819–1829. PMID: 16737951.9. Wulfkuhle JD, Paweletz CP, Steeg PS, Petricoin EF 3rd, Liotta L. Proteomic approaches to the diagnosis, treatment, and monitoring of cancer. Adv Exp Med Biol. 2003; 532:59–68. PMID: 12908550.
Article10. Bichsel VE, Liotta LA, Petricoin EF 3rd. Cancer proteomics: from biomarker discovery to signal pathway profiling. Cancer J. 2001; 7:69–78. PMID: 11269650.11. Verma M, Wright GL Jr, Hanash SM, Gopal-Srivastava R, Srivastava S. Proteomic approaches within the NCI early detection research network for the discovery and identification of cancer biomarkers. Ann N Y Acad Sci. 2001; 945:103–115. PMID: 11708463.
Article12. Stauber J, MacAleese L, Franck J, et al. On-tissue protein identification and imaging by MALDI-ion mobility mass spectrometry. J Am Soc Mass Spectrom. 2010; 21:338–347. PMID: 19926301.
Article13. Schwamborn K. Imaging mass spectrometry in biomarker discovery and validation. J Proteomics. 2012; 75:4990–4998. PMID: 22749859.
Article14. MacAleese L, Stauber J, Heeren RM. Perspectives for imaging mass spectrometry in the proteomics landscape. Proteomics. 2009; 9:819–834. PMID: 19212956.
Article15. Ye H, Gemperline E, Li L. A vision for better health: mass spectrometry imaging for clinical diagnostics. Clin Chim Acta. 2013; 420:11–22. PMID: 23078851.
Article16. Passarelli MK, Ewing AG. Single-cell imaging mass spectrometry. Curr Opin Chem Biol. Epub 2013 Aug 12. DOI: 10.1016/j.cbpa.2013.07.017.
Article17. Grüner BM, Hahne H, Mazur PK, et al. MALDI imaging mass spectrometry for in situ proteomic analysis of preneoplastic lesions in pancreatic cancer. PLoS One. 2012; 7:e39424. PMID: 22761793.
Article18. Meding S, Walch A. MALDI imaging mass spectrometry for direct tissue analysis. Methods Mol Biol. 2013; 931:537–546. PMID: 23027023.
Article19. Zoehrer R, Perilli E, Kuliwaba JS, Shapter JG, Fazzalari NL, Voelcker NH. Human bone material characterization: integrated imaging surface investigation of male fragility fractures. Osteoporos Int. 2012; 23:1297–1309. PMID: 21695535.
Article20. Klerk LA, Dankers PY, Popa ER, et al. TOF-secondary ion mass spectrometry imaging of polymeric scaffolds with surrounding tissue after in vivo implantation. Anal Chem. 2010; 82:4337–4343. PMID: 20462187.
Article21. Tanaka H, Zaima N, Yamamoto N, et al. Distribution of phospholipid molecular species in autogenous access grafts for hemodialysis analyzed using imaging mass spectrometry. Anal Bioanal Chem. 2011; 400:1873–1880. PMID: 21404091.
Article22. Thiele H, Heldmann S, Trede D, et al. 2D and 3D MALDI-imaging: conceptual strategies for visualization and data mining. Biochim Biophys Acta. 2013; 3. 04. Epub. DOI: 10.1016/j.bbapap.2013.01.040.
Article23. Andersson M, Groseclose MR, Deutch AY, Caprioli RM. Imaging mass spectrometry of proteins and peptides: 3D volume reconstruction. Nat Methods. 2008; 5:101–108. PMID: 18165806.
Article24. Lemaire R, Desmons A, Tabet JC, Day R, Salzet M, Fournier I. Direct analysis and MALDI imaging of formalin-fixed, paraffin-embedded tissue sections. J Proteome Res. 2007; 6:1295–1305. PMID: 17291023.
Article25. Casadonte R, Caprioli RM. Proteomic analysis of formalin-fixed paraffin-embedded tissue by MALDI imaging mass spectrometry. Nat Protoc. 2011; 6:1695–1709. PMID: 22011652.
Article26. Takai N, Tanaka Y, Inazawa K, Saji H. Quantitative analysis of pharmaceutical drug distribution in multiple organs by imaging mass spectrometry. Rapid Commun Mass Spectrom. 2012; 26:1549–1556. PMID: 22638972.
Article27. Takai N, Tanaka Y, Watanabe A, Saji H. Quantitative imaging of a therapeutic peptide in biological tissue sections by MALDI MS. Bioanalysis. 2013; 5:603–612. PMID: 23425275.
Article28. Terp MG, Lund RR, Jensen ON, Leth-Larsen R, Ditzel HJ. Identification of markers associated with highly aggressive metastatic phenotypes using quantitative comparative proteomics. Cancer Genomics Proteomics. 2012; 9:265–273. PMID: 22990106.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Risk of inaccurate species identification by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and of false carbapenem resistance by automated susceptibility analysis of Enterobacter spp.
- Reliability of Acinetobacter baumannii Identification with Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry
- Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry in Clinical Microbiology: What Are the Current Issues?
- MALDI-TOF-MS Fingerprinting Provides Evidence of Urosepsis caused by Aerococcus urinae
- Evaluation of two commercial kits for rapid pathogen identification directly from positive blood cultures by matrix-associated laser desorption/ionization time-of-flight mass spectrometry