Immune Netw.
2009 Dec;9(6):243-247. 10.4110/in.2009.9.6.243.
A New Reporter Vector System Based on Flow-Cytometry to Detect Promoter Activity
- Affiliations
-
- 1School of Life Sciences and Biotechnology, Korea University, Seoul 136-701, Korea. sehopark@korea.ac.kr
- 2Department of Microbiology, Gachon Medical School, Inchon 405-760, Korea.
- KMID: 2150652
- DOI: http://doi.org/10.4110/in.2009.9.6.243
Abstract
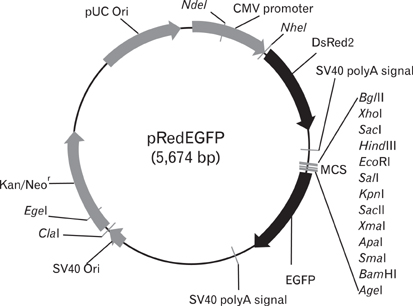
- In this study, we report the development of a new dual reporter vector system for the analysis of promoter activity. This system employs green fluorescence emitting protein, EGFP, as a reporter, and uses red fluorescence emitting protein, DsRed, as a transfection control in a single vector. The expression of those two proteins can be readily detected via flow cytometry in a single analysis, with no need for any further manipulation after transfection. As this system allows for the simultaneous detection of both the control and reporter proteins in the same cells, only transfected cells which express the control protein, DsRed, can be subjected to promoter activity analysis, via the gating out of all un-transfected cells. This results in a dramatic increase in the promoter activity detection sensitivity. This novel reporter vector system should prove to be a simple and efficient method for the analysis of promoter activity.
MeSH Terms
Figure
Reference
-
1. Dominguez P, Ibaraki K, Robey PG, Hefferan TE, Termine JD, Young MF. Expression of the osteonectin gene potentially controlled by multiple cis- and trans-acting factors in cultured bone cells. J Bone Miner Res. 1991. 6:1127–1136.
Article2. Gorman CM, Moffat LF, Howard BH. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982. 2:1044–1051.
Article3. Alam J, Cook JL. Reporter genes: application to the study of mammalian gene transcription. Anal Biochem. 1990. 188:245–254.
Article4. Russo-Marie F, Roederer M, Sager B, Herzenberg LA, Kaiser D. Beta-galactosidase activity in single differentiating bacterial cells. Proc Natl Acad Sci U S A. 1993. 90:8194–8198.
Article5. Dietrich C, Maiss E. Red fluorescent protein DsRed from Discosoma sp. as a reporter protein in higher plants. Biotechniques. 2002. 32:286–293.
Article6. Nancharaiah YV, Wattiau P, Wuertz S, Bathe S, Mohan SV, Wilderer PA, Hausner M. Dual labeling of Pseudomonas putida with fluorescent proteins for in situ monitoring of conjugal transfer of the TOL plasmid. Appl Environ Microbiol. 2003. 69:4846–4852.
Article7. Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene. 1996. 173(1 Spec No):33–38.
Article8. Gross LA, Baird GS, Hoffman RC, Baldridge KK, Tsien RY. The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci U S A. 2000. 97:11990–11995.
Article9. Davey HM, Kell DB. Flow cytometry and cell sorting of heterogeneous microbial populations: the importance of single-cell analyses. Microbiol Rev. 1996. 60:641–696.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Heterologous Regulation of BCG hsp65 Promoter by M. leprae 18 kDa Transcription Repression Responsive Element
- Development of an efficient endothelial cell specific vector using promoter and 5' untranslated sequences from the human preproendothelin-1 gene
- Recombinant AAV Vector with MITF-M Promoter for Melanoma Gene Therapy
- Analysis of T Cells Using Flow Cytometry
- A Novel Rapid Fungal Promoter Analysis System Using the Phosphopantetheinyl Transferase Gene, npgA, in Aspergillus nidulans