Ann Lab Med.
2014 May;34(3):240-242. 10.3343/alm.2014.34.3.240.
Analysis of Reporting Time for Identification of Methicillin-Resistant Staphylococcus aureus Carriers Using ChromID MRSA
- Affiliations
-
- 1Department of Laboratory Medicine, Hallym University College of Medicine, Kangdong Sacred Heart Hospital, Seoul, Korea. jaeseok@hallym.or.kr
- KMID: 1791927
- DOI: http://doi.org/10.3343/alm.2014.34.3.240
Abstract
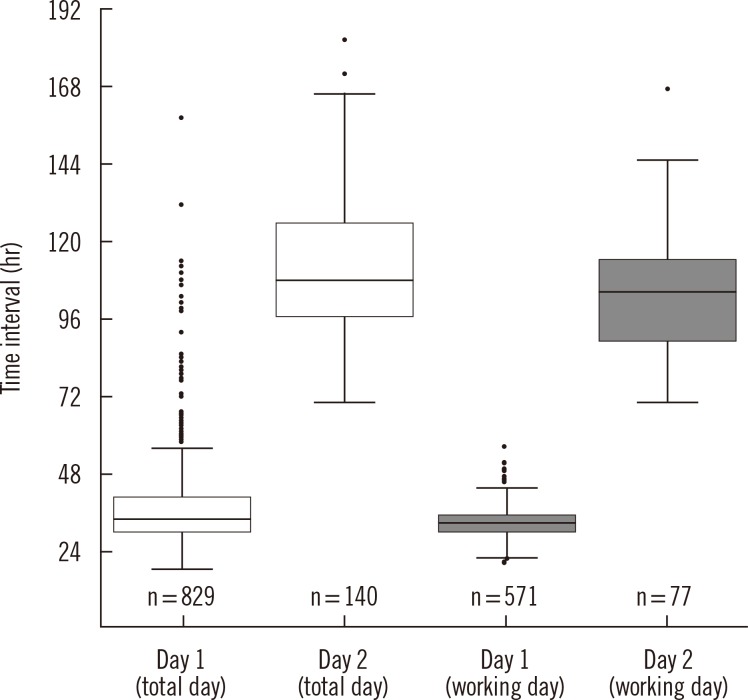
- We assessed the reporting times for identification of nasal methicillin-resistant Staphylococcus aureus (MRSA) carriers in 2011 in a university-affiliated hospital using surveillance cultures incubated for 1 and 2 days with ChromID MRSA (bioMerieux, France). Of 2,732 nasal swabs tested, MRSA was detected in 829 (85.6%) and 140 (14.4%) swabs after 1 and 2 days of incubation, respectively, and the median reporting times for positive specimens were 33.7 hr (range, 18.2-156.9 hr) and 108.1 hr (range, 69.8-181.0 hr), respectively. Detection rate after 1-day incubation was 85%. Additional 1-day incubation improved detection rate; however, it prolonged the reporting times of positive specimens approximately up to 4 days because of the need for confirmatory tests such as species identification and susceptibility tests. Following a 2-day culture with ChromID MRSA, rapid confirmatory tests are warranted to reduce delay in identifying MRSA carriers.
Keyword
MeSH Terms
Figure
Reference
-
1. Cosgrove SE, Sakoulas G, Perencevich EN, Schwaber MJ, Karchmer AW, Carmeli Y. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin Infect Dis. 2003; 36:53–59. PMID: 12491202.2. Engemann JJ, Carmeli Y, Cosgrove SE, Fowler VG, Bronstein MZ, Trivette SL, et al. Adverse clinical and economic outcomes attributable to methicillin resistance among patients with Staphylococcus aureus surgical site infection. Clin Infect Dis. 2003; 36:592–598. PMID: 12594640.3. Lee H, Kim CK, Lee J, Lee SH, Ahn JY, Hong SG, et al. Antimicrobial resistance of clinically important bacteria isolated from 12 hospitals in Korea in 2005 and 2006. Korean J Clin Microbiol. 2007; 10:59–69.4. Tacconelli E, De Angelis G, de Waure C, Cataldo MA, La Torre G, Cauda R. Rapid screening tests for meticillin-resistant Staphylococcus aureus at hospital admission: systematic review and meta-analysis. Lancet Infect Dis. 2009; 9:546–554. PMID: 19695491.5. Harbarth S, Hawkey PM, Tenover F, Stefani S, Pantosti A, Struelens MJ. Update on screening and clinical diagnosis of meticillin-resistant Staphylococcus aureus (MRSA). Int J Antimicrob Agents. 2011; 37:110–117. PMID: 21163628.6. Malhotra-Kumar S, Haccuria K, Michiels M, Ieven M, Poyart C, Hryniewicz W, et al. Current trends in rapid diagnostics for methicillin-resistant Staphylococcus aureus and glycopeptide-resistant enterococcus species. J Clin Microbiol. 2008; 46:1577–1587. PMID: 18322065.7. Wassenberg MW, Kluytmans JA, Box AT, Bosboom RW, Buiting AG, van Elzakker EP, et al. Rapid screening of methicillin-resistant Staphylococcus aureus using PCR and chromogenic agar: a prospective study to evaluate costs and effects. Clin Microbiol Infect. 2010; 16:1754–1761. PMID: 20219077.8. Morris K, Wilson C, Wilcox MH. Evaluation of chromogenic meticillin-resistant Staphylococcus aureus media: sensitivity versus turnaround time. J Hosp Infect. 2012; 81:20–24. PMID: 22463978.9. Carson J, Lui B, Rosmus L, Rennick H, Fuller J. Interpretation of MRSASelect screening agar at 24 hours of incubation. J Clin Microbiol. 2009; 47:566–568. PMID: 19144795.10. Nahimana I, Francioli P, Blanc DS. Evaluation of three chromogenic media (MRSA-ID, MRSA-Select and CHROMagar MRSA) and ORSAB for surveillance cultures of methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2006; 12:1168–1174. PMID: 17121622.11. Nonhoff C, Denis O, Brenner A, Buidin P, Legros N, Thiroux C, et al. Comparison of three chromogenic media and enrichment broth media for the detection of methicillin-resistant Staphylococcus aureus from mucocutaneous screening specimens: comparison of MRSA chromogenic media. Eur J Clin Microbiol Infect Dis. 2009; 28:363–369. PMID: 18855028.12. Van Hoecke F, Deloof N, Claeys G. Performance evaluation of a modified chromogenic medium, ChromID MRSA New, for the detection of methicillin-resistant Staphylococcus aureus from clinical specimens. Eur J Clin Microbiol Infect Dis. 2011; 30:1595–1598. PMID: 21499707.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Evaluation of ChromID MRSA for the Detection of Methicillin-resistant Staphylococcus aureus
- Detection of Multidrug Resistant Patterns and Associated - genes of Methicillin Resistant Staphylococcus aureus ( MRSA ) Isolated from Clinical Specimens
- A case of multiple furunculosis caused by methicillin-resistant staphylococcs aureus
- Community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA)
- Molecular Epidemiologic Methods Used in the Analysis of Methicillin-Resistant staphylococcus aureus