Immune Netw.
2009 Aug;9(4):138-146. 10.4110/in.2009.9.4.138.
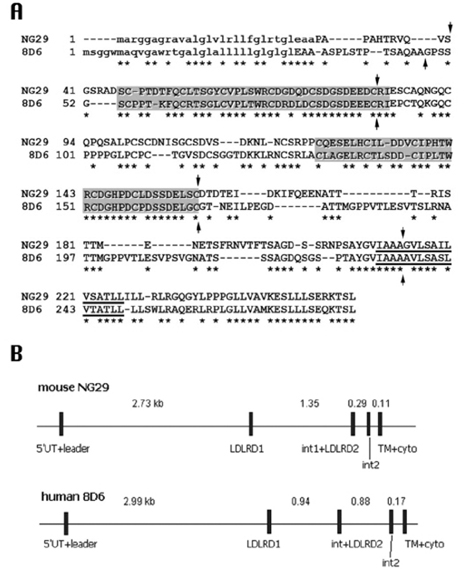
Characterization of a Novel Gene in the Extended MHC Region of Mouse, NG29/Cd320, a Homolog of the Human CD320
- Affiliations
-
- 1Department of Pathology, Seoul National University College of Medicine, Seoul, Korea.
- 2Division of Immunology, Department of Molecular Cell Biology, Samsung Biomedical Research Institute, Sungkyunkwan University School of Medicine, Suwon, Korea. tjkim@med.skku.ac.kr
- KMID: 1474586
- DOI: http://doi.org/10.4110/in.2009.9.4.138
Abstract
- BACKGROUND
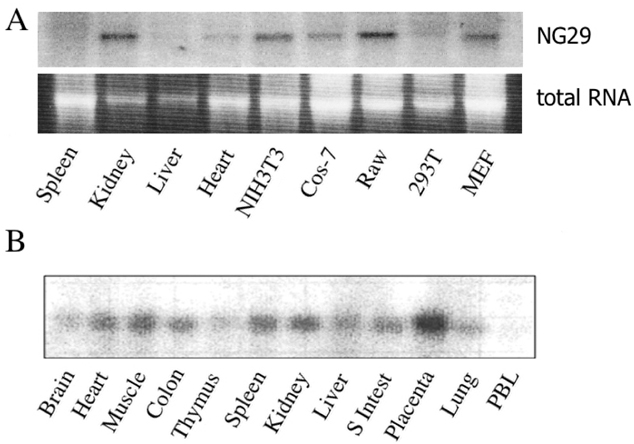
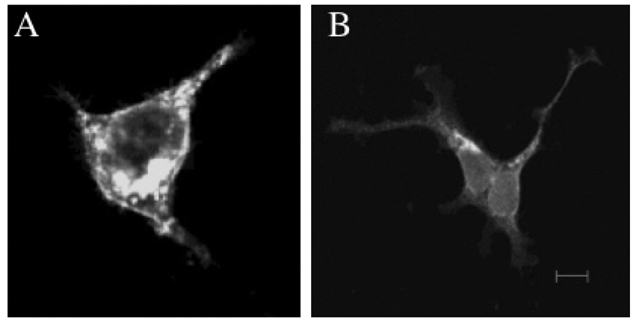
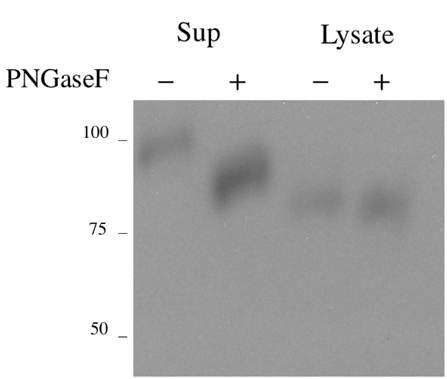
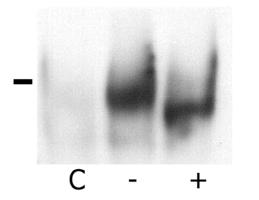
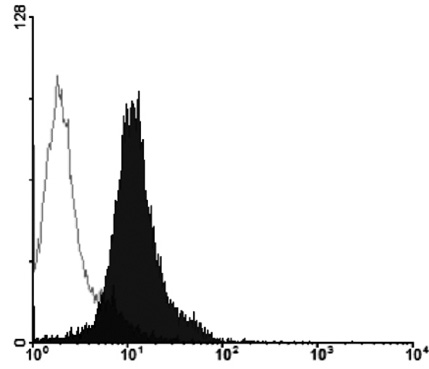
The MHC region of the chromosome contains a lot of genes involved in immune responses. Here we have investigated the mouse NG29/Cd320 gene in the centrometrically extended MHC region of chromosome 17. METHODS: We cloned the NG29 gene by RT-PCR and confirmed the tissue distribution of its gene expression by northern blot hybridization. We generated the NG29 gene expression constructs and polyclonal antibody against the NG29 protein to perform the immunofluorescence, immunoprecipitation and flow cytometric analysis. RESULTS: The murine NG29 gene and its human homologue, the CD320/8D6 gene, were similar in the gene structure and tissue expression patterns. We cloned the NG29 gene and confirmed its expression in plasma membrane and intracellular compartments by transfecting its expresssion constructs into HEK 293T cells. The immunoprecipitation studies with rabbit polyclonal antibody raised against the NG29-NusA fusion protein indicated that NG29 protein was a glycoprotein of about 45 kDa size. A flow cytometric analysis also showed the NG29 expression on the surface of Raw 264.7 macrophage cell line. CONCLUSION: These findings suggested that NG29 gene in mouse extended MHC class II region was the orthologue of human CD320 gene even though human CD320/8D6 gene was located in non-MHC region, chromosome 19p13.
Keyword
MeSH Terms
Figure
Reference
-
1. Amadou C, Kumánovics A, Jones EP, Lambracht-Washington D, Yoshino M, Lindahl KF. The mouse major histocompatibility complex: some assembly required. Immunol Rev. 1999. 167:211–221.
Article2. Yeager M, Hughes AL. Evolution of the mammalian MHC: natural selection, recombination, and convergent evolution. Immunol Rev. 1999. 167:45–58.
Article3. Stephens R, Horton R, Humphray S, Rowen L, Trowsdale J, Beck S. Gene organisation, sequence variation and isochore structure at the centromeric boundary of the human MHC. J Mol Biol. 1999. 291:789–799.
Article4. Gruen JR, Weissman SM. Evolving views of the major histocompatibility complex. Blood. 1997. 90:4252–4265.
Article5. Complete sequence and gene map of a human major histocompatibility complex. The MHC sequencing consortium. Nature. 1999. 401:921–923.6. Hauptmann G, Bahram S. Genetics of the central MHC. Curr Opin Immunol. 2004. 16:668–672.
Article7. Cupedo T, Kraal G, Mebius RE. The role of CD45+CD4+CD3- cells in lymphoid organ development. Immunol Rev. 2002. 189:41–50.8. Anderson G, Jenkinson EJ. Lymphostromal interactions in thymic development and function. Nat Rev Immunol. 2001. 1:31–40.
Article9. Li L, Choi Y. Follicular dendritic cell-signaling molecules required for proliferation and differentiation of GC-B cells. Semin Immunol. 2002. 14:259–266.
Article10. Katakai T, Hara T, Sugai M, Gonda H, Shimizu A. Lymph node fibroblastic reticular cells construct the stromal reticulum via contact with lymphocytes. J Exp Med. 2004. 200:783–795.
Article11. Gretz JE, Anderson AO, Shaw S. Cords, channels, corridors and conduits: critical architectural elements facilitating cell interactions in the lymph node cortex. Immunol Rev. 1997. 156:11–24.
Article12. Bofill M, Akbar AN, Amlot PL. Follicular dendritic cells share a membrane-bound protein with fibroblasts. J Pathol. 2000. 191:217–226.
Article13. Balogh P, Horváth G, Szakal AK. Immunoarchitecture of distinct reticular fibroblastic domains in the white pulp of mouse spleen. J Histochem Cytochem. 2004. 52:1287–1298.
Article14. Li L, Zhang X, Kovacic S, Long AJ, Bourque K, Wood CR, Choi YS. Identification of a human follicular dendritic cell molecule that stimulates germinal center B cell growth. J Exp Med. 2000. 191:1077–1084.
Article15. Zhang X, Li L, Jung J, Xiang S, Hollmann C, Choi YS. The distinct roles of T cell-derived cytokines and a novel follicular dendritic cell-signaling molecule 8D6 in germinal center-B cell differentiation. J Immunol. 2001. 167:49–56.
Article16. Kim JY, Lee MH, Jung KI, Na HY, Cha HS, Ko EM, Kim TJ. Detection of antibodies against glucose 6-phosphate isomerase in synovial fluid of rheumatoid arthritis using surface plasmon resonance (BIAcore). Exp Mol Med. 2003. 35:310–316.
Article17. Hahn MJ, Yoon SS, Sohn HW, Song HG, Park SH, Kim TJ. Differential activation of MAP kinase family members triggered by CD99 engagement. FEBS Lett. 2000. 470:350–354.
Article18. Herz J, Hamann U, Rogne S, Myklebost O, Gausepohl H, Stanley KK. Surface location and high affinity for calcium of a 500-kd liver membrane protein closely related to the LDL-receptor suggest a physiological role as lipoprotein receptor. EMBO J. 1988. 7:4119–4127.
Article19. Krieger M, Herz J. Structures and functions of multiligand lipoprotein receptors: macrophage scavenger receptors and LDL receptor-related protein (LRP). Annu Rev Biochem. 1994. 63:601–637.
Article20. Bar I, Goffinet AM. Developmental neurobiology. Decoding the Reelin signal. Nature. 1999. 399:645–646.21. Trommsdorff M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, Hammer RE, Richardson JA, Herz J. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999. 97:689–701.
Article22. Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999. 1473:4–8.
Article23. Earl LA, Baum LG. CD45 glycosylation controls T-cell life and death. Immunol Cell Biol. 2008. 86:608–615.
Article24. Zhao Y, Sato Y, Isaji T, Fukuda T, Matsumoto A, Miyoshi E, Gu J, Taniguchi N. Branched N-glycans regulate the biological functions of integrins and cadherins. FEBS J. 2008. 275:1939–1948.
Article25. Kasahara M, Hayashi M, Tanaka K, Inoko H, Sugaya K, Ikemura T, Ishibashi T. Chromosomal localization of the proteasome Z subunit gene reveals an ancient chromosomal duplication involving the major histocompatibility complex. Proc Natl Acad Sci U S A. 1996. 93:9096–9101.
Article26. Kasahara M, Nakaya J, Satta Y, Takahata N. Chromosomal duplication and the emergence of the adaptive immune system. Trends Genet. 1997. 13:90–92.
Article27. Katsanis N, Fitzgibbon J, Fisher EM. Paralogy mapping: identification of a region in the human MHC triplicated onto human chromosomes 1 and 9 allows the prediction and isolation of novel PBX and NOTCH loci. Genomics. 1996. 35:101–108.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Species specificity of a novel factor which augments the expression of MHC class I antigens on tumor cell lines
- Transcription of class II MHC gene by interferon-gamma in FRTL-5 cells
- Identification and Functional Characterization of a Cryptococcus neoformans UPC2 Homolog
- Nuclear protein binding patterns in the 5'-upstream regulatory elements of HLA class I genes
- Human Embryonic Stem Cell Derived from Early Stage Fertilized Ovum: Non Immunogenic and Universal, Neuronal and Non-neuronal Cell Lines