J Vet Sci.
2012 Sep;13(3):271-278. 10.4142/jvs.2012.13.3.271.
Frequency of group A rotavirus with mixed G and P genotypes in bovines: predominance of G3 genotype and its emergence in combination with G8/G10 types
- Affiliations
-
- 1Division of Virology, Indian Veterinary Research Institute, Campus Mukteswar 263138, Uttarakhand, India. malikyps@yahoo.com
- 2College of Veterinary Sciences, Jawaharlal Nehru Krishi Vishwa Vidyalaya, Jabalpur, Madhya Pradesh 428001, India.
- 3GB Pant University of Agriculture and Technology, Pantnagar 263145, Uttarakhand, India.
- 4Department of Animal Biotechnology, College of Veterinary Sciences, CCS Haryana Agricultural University, Hisar 125004, Haryana, India.
- 5National Research Centre on Equine, Sirsa Road, Hisar 125001, Haryana, India.
- 6Division of Veterinary Public Health, Indian Veterinary Research Institute, Izatnagar 243122, Utter Pradesh, India.
- KMID: 1389766
- DOI: http://doi.org/10.4142/jvs.2012.13.3.271
Abstract
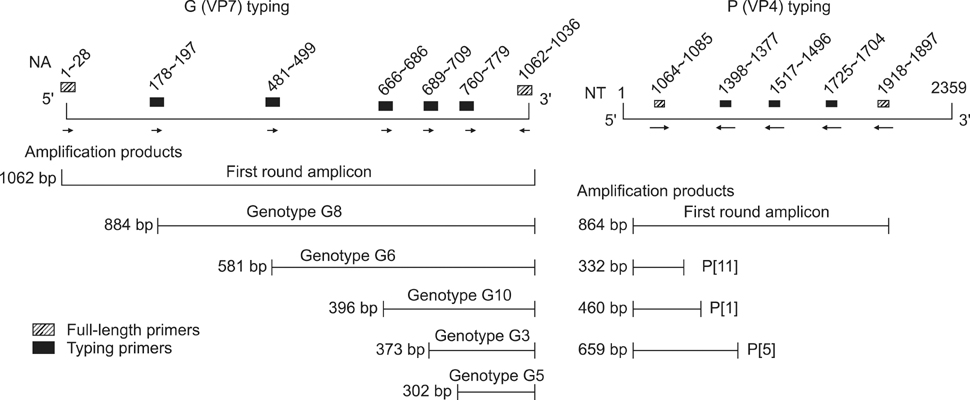
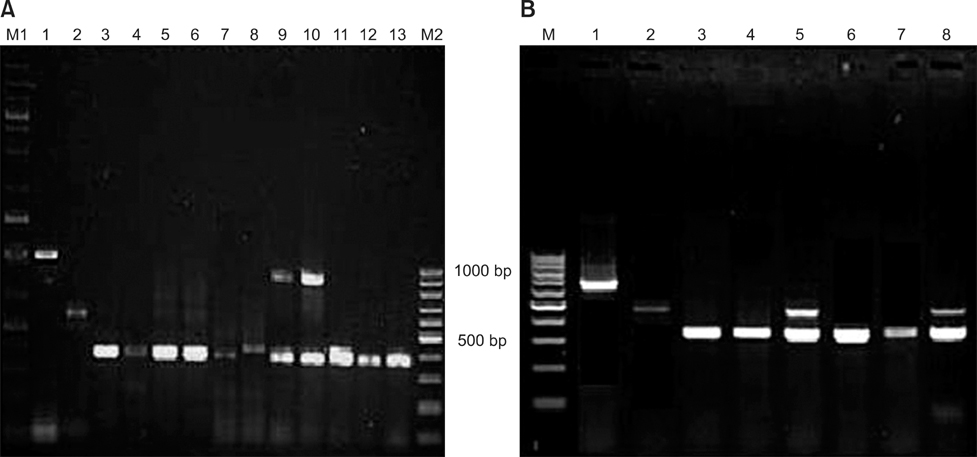
- The present study describes the genotypic distribution of rotaviruses (RVs) in an Indian bovine population with unexpectedly higher proportions of G3 alone or in combination of G8/G10. PCR-genotyping confirmed that 39.4% (13/33) of the prevalent RVs were the G3 type while 60.6% (20/33) were dual G3G10 or G3G8 types. P typing revealed that 93.9% (31/33) of the samples were P[11] while 6.1% (2/33) possessed a dual P[1]P[11] type. Sequence analysis of the VP7 gene from G3 strains viz. B-46, 0970, and BR-133 showed that these strains had sequence identities of 90.5% to 100% with other bovine G3 strains. The highest identity (98.9% to 100%) was observed with RUBV3 bovine G3 strains from eastern India. The G3 strains (B-46, 0970, and BR-133) showed 97.5% to 98.8% sequence homologies with the Indian equine RV strain Erv-80. Phylogenetic analysis demonstrated that G3 strains clustered with bovine RUBV3 and J-63, and equine Erv-80 G3. Overall, these results confirmed that the incidence of infection by RVs with the G3 genotype and mixed genotypes in the bovine population was higher than previously predicted. This finding reinforces the importance of constantly monitoring circulating viral strains with the G3 genotype in future surveillance studies.
MeSH Terms
-
Animals
Cattle
Cattle Diseases/epidemiology/*virology
Desert Climate
Feces/virology
Genotype
India/epidemiology
Molecular Sequence Data
Phylogeny
RNA, Viral/genetics
Reverse Transcriptase Polymerase Chain Reaction/veterinary
Rotavirus/classification/*genetics/isolation & purification
Rotavirus Infections/epidemiology/*veterinary/virology
Sequence Analysis, Protein/veterinary
Sequence Analysis, RNA/veterinary
Sequence Homology
Tropical Climate
Figure
Reference
-
1. Aladin F, Nawaz S, Iturriza-Gómara M, Gray J. Identification of G8 rotavirus strains determined as G12 by rotavirus genotyping PCR: updating the current genotyping methods. J Clin Virol. 2010. 47:340–344.
Article2. Cashman O, Lennon G, Sleator RD, Power E, Fanning S, O'Shea H. Changing profile of the bovine rotavirus G6 population in the south of Ireland from 2002 to 2009. Vet Microbiol. 2010. 146:238–244.
Article3. Collins PJ, Martella V, Buonavoglia C, O'Shea H. Identification of a G2-like porcine rotavirus bearing a novel VP4 type, P[32]. Vet Res. 2010. 41:73.
Article4. Das BK, Gentsch JR, Cicirello HG, Woods PA, Gupta A, Ramachandran M, Kumar R, Bhan MK, Glass RI. Characterization of rotavirus strains from newborns in New Delhi, India. J Clin Microbiol. 1994. 32:1820–1822.
Article5. El-Attar L, Dhaliwal W, Iturriza-Gómara M, Bridger JC. Identification and molecular characterization of a bovine G3 rotavirus which causes age-independent diarrhea in cattle. J Clin Microbiol. 2002. 40:937–942.
Article6. Esona MD, Mijatovic-Rustempasic S, Conrardy C, Tong S, Kuzmin IV, Agwanda B, Breiman RF, Banyai K, Niezgoda M, Rupprecht CE, Gentsch JR, Bowen MD. Reassortant group A rotavirus from straw-colored fruit bat (Eidolon helvum). Emerg Infect Dis. 2010. 16:1844–1852.
Article7. Estes MK, Kapikian AZ. Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Rotaviruses. Fields' Virology. 2007. Vol. 2:5th ed. Philadelphia: Lippincott Williams & Wilkins;1917–1974.8. Fagiolo A, Roncoroni C, Lai O, Borghese A. Borghese A, editor. Buffalo pathologies. Buffalo Production and Research. 2005. Rome: FAO;249–296.9. Fukai K, Maeda Y, Fujimoto K, Itou T, Sakai T. Changes in the prevalence of rotavirus G and P types in diarrheic calves from the Kagoshima prefecture in Japan. Vet Microbiol. 2002. 86:343–349.
Article10. Ghosh S, Samajdar S, Sinha M, Kobayashi N, Taniguchi K, Naik TN. Molecular characterization of rare bovine group A rotavirus G15P[11] and G15P[21] strains from eastern India: identification of simian SA11-like VP6 genes in G15P[21] strains. Virus Genes. 2008. 37:241–249.
Article11. Ghosh S, Varghese V, Samajdar S, Sinha M, Kobayashi N, Naik TN. Molecular characterization of bovine group A rotavirus G3P[3] strains. Arch Virol. 2007. 152:1935–1940.
Article12. Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, Forrester B, Fang ZY. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol. 1990. 28:276–282.
Article13. Gouvea V, Santos N, Timenetsky Mdo C. Identification of bovine and porcine rotavirus G types by PCR. J Clin Microbiol. 1994. 32:1338–1340.
Article14. Gulati BR, Nakagomi O, Koshimura Y, Nakagomi T, Pandey R. Relative frequencies of G and P types among rotaviruses from Indian diarrheic cow and buffalo calves. J Clin Microbiol. 1999. 37:2074–2076.
Article15. Isegawa Y, Nakagomi O, Nakagomi T, Ishida S, Uesugi S, Ueda S. Determination of bovine rotavirus G and P serotypes by polymerase chain reaction. Mol Cell Probes. 1993. 7:277–284.
Article16. Iturriza-Gómara M, Kang G, Gray J. Rotavirus genotyping: keeping up with an evolving population of human rotaviruses. J Clin Virol. 2004. 31:259–265.
Article17. Kapikian AZ, Hoshino Y, Chanock RM, Pérez-Schael I. Efficacy of a quadrivalent rhesus rotavirus-based human rotavirus vaccine aimed at preventing severe rotavirus diarrhea in infants and young children. J Infect Dis. 1996. 174:Suppl 1. S65–S72.
Article18. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970. 227:680–685.
Article19. Maes P, Matthijnssens J, Rahman M, Van Ranst M. RotaC: a web-based tool for the complete genome classification of group A rotaviruses. BMC Microbiol. 2009. 9:238.
Article20. Manuja BK, Prasad M, Manuja A, Gulati BR, Prasad G. A novel genomic constellation (G10P[3]) of group A rotavirus detected from buffalo calves in northern India. Virus Res. 2008. 138:36–42.
Article21. Matthijnssens J, Rahman M, Ciarlet M, Zeller M, Heylen E, Nakagomi T, Uchida R, Hassan Z, Azim T, Nakagomi O, Van Ranst M. Reassortment of human rotavirus gene segments into G11 rotavirus strains. Emerg Infect Dis. 2010. 16:625–630.
Article22. Minakshi , Prasad G, Malik Y, Pandey R. G and P genotyping of bovine group A rotaviruses in faecal samples of diarrhoeic calves by DIG-labelled probes. Indian J Biotechnol. 2005. 4:93–99.23. Okada N, Matsumoto Y. Bovine rotavirus G and P types and sequence analysis of the VP7 gene of two G8 bovine rotaviruses from Japan. Vet Microbiol. 2002. 84:297–305.
Article24. Parra GI, Espinola EE. Nucleotide mismatches between the VP7 gene and the primer are associated with genotyping failure of a specific lineage from G1 rotavirus strains. Virol J. 2006. 3:35.25. Rao CD, Gowda K, Reddy BSY. Sequence analysis of VP4 and VP7 genes of nontypeable strains identifies a new pair of outer capsid proteins representing novel P and G genotypes in bovine rotaviruses. Virology. 2000. 276:104–113.
Article26. Reidy N, Lennon G, Fanning S, Power E, O'Shea H. Molecular characterization and analysis of bovine rotavirus strains circulating in Ireland 2002-2004. Vet Microbiol. 2006. 117:242–247.
Article27. Saravanan M, Parthiban M, Ramadass P. Genotyping of rotavirus of neonatal calves by nested-multiplex PCR in India. Vet Arhiv. 2006. 76:497–505.28. Sharma G, Taku A, Chhabra R, Bhat MA. Prevalence of bovine rotaviral diarrhoea in Jammu. Indian J Virol. 2009. 20:53–58.29. Svensson L, Uhnoo I, Grandien M, Wadell G. Molecular epidemiology of rotavirus infections in Uppsala, Sweden, 1981: disappearance of a predominant electropherotype. J Med Virol. 1986. 18:101–111.
Article30. Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007. 24:1596–1599.
Article31. Taniguchi K, Wakasugi F, Pongsuwanna Y, Urasawa T, Ukae S, Chiba S, Urasawa S. Identification of human and bovine rotavirus serotypes by polymerase chain reaction. Epidemiol Infect. 1992. 109:303–312.
Article32. Ursu K, Kisfali P, Rigó D, Ivanics E, Erdélyi K, Dán A, Melegh B, Martella V, Bányai K. Molecular analysis of the VP7 gene of pheasant rotaviruses identifies a new genotype, designated G23. Arch Virol. 2009. 154:1365–1369.
Article33. Varshney B, Jagannath MR, Vethanayagam RR, Kodhandharaman S, Jagannath HV, Gowda K, Singh DK, Rao CD. Prevalence of, and antigenic variation in, serotype G10 rotaviruses and detection of serotype G3 strains in diarrheic calves: implications for the origin of G10P11 or P11 type reassortant asymptomatic strains in newborn children in India. Arch Virol. 2002. 147:143–165.
Article34. Wani SA, Bhat MA, Ishaq SM, Ashrafi MA. Determination of bovine rotavirus G genotypes in Kashmir, India. Rev Sci Tech. 2004. 23:931–936.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- VP7 Genotypes of Group A Rotavirus Isolated from Infants and Toddlers with Rotavirus Gastroenteritis in Jeju
- VP4 and VP7 Genotyping of Group A Rotavirus Isolated from Diarrhea Patients in Seoul by Multiplex PCR
- Distribution of Human Rotavirus Genotypes in a Tertiary Hospital, Seoul, Korea During 2009-2013
- Rotavirus P and G Genotypes Circulating in Kyungsangnamdo, Korea, during 2000~2001
- Prevalence of rotavirus genotypes in South Korea in 1989-2009: implications for a nationwide rotavirus vaccine program