J Korean Med Sci.
2008 Dec;23(6):1053-1061. 10.3346/jkms.2008.23.6.1053.
Comparison of Her-2, EGFR and Cyclin D1 in Primary Breast Cancer and Paired Metastatic Lymph Nodes: An Immunohistochemical and Chromogenic In Situ Hybridization Study
- Affiliations
-
- 1Department of Pathology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea. bijou@skku.edu
- KMID: 1107533
- DOI: http://doi.org/10.3346/jkms.2008.23.6.1053
Abstract
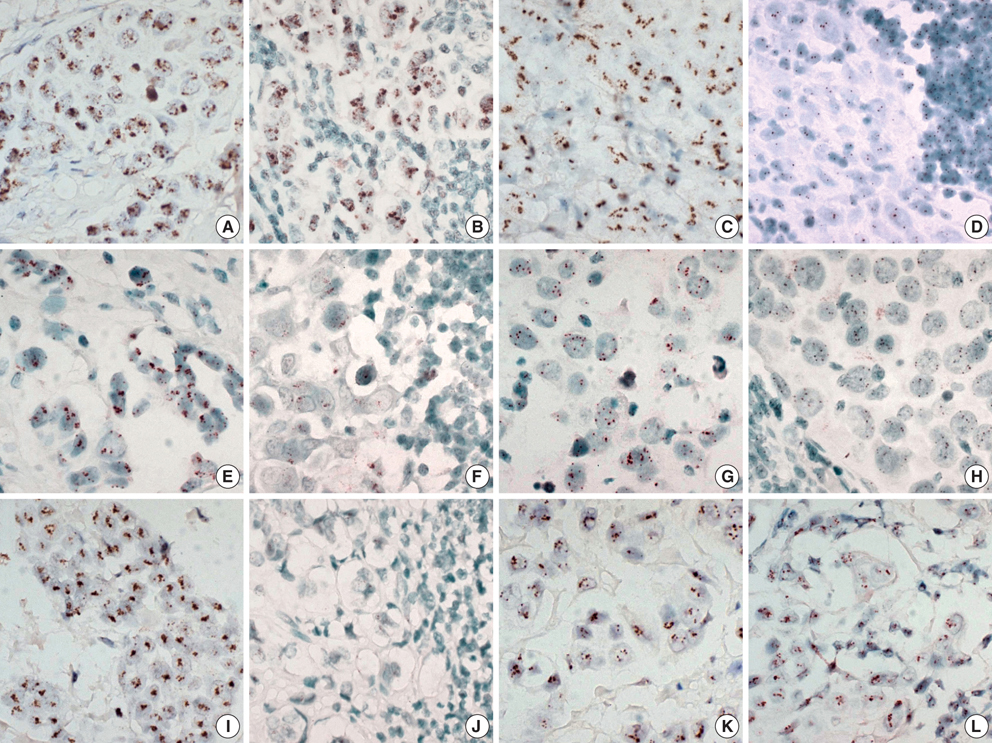
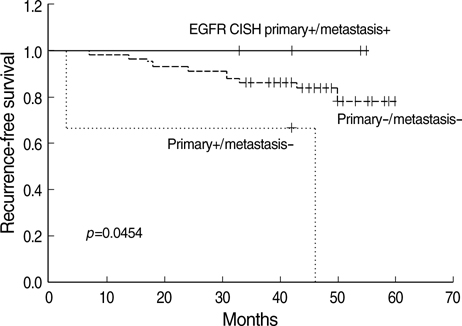
- The significant advance in the development of molecular-targeting drugs has made an evaluation of Her-2, EGFR, and cyclin D1 an important clinical issue in breast cancer patients. This study compared the Her-2, EGFR, and cyclin D1 status of primary tumors as well as their matching lymph node metastases using immunohistochemistry (IHC) and chromogenic in situ hybridization (CISH) in 73 breast cancer patients. Her-2, EGFR, and cyclin D1 protein showed a concordance between the primary lesion and the metastatic regional lymph nodes in 82%, 90%, and 63%, respectively. CISH also revealed 92%, 93%, and 85% concordance in the gene amplification status of Her-2, EGFR, and cyclin D1, showing a reasonable agreement between primary tumors and metastatic regional lymph nodes. Although a statistically significant agreement was found in Her-2 expression, a relatively high discordance rate (18%) raises a little concern. Our findings suggest that the Her-2 status can be reliably assessed on primary tumor but a possible difference can be found in Her-2, EGFR, and cyclin D1 status between the primary and the metastatic sites and this possibility should be concerned in patients considering molecular targeted therapy or patients with progress of disease.
Keyword
MeSH Terms
-
Adult
Aged
Breast Neoplasms/genetics/*metabolism/pathology
Chromogenic Compounds
Cyclin D1/*analysis/genetics
Female
Humans
Immunohistochemistry
In Situ Hybridization
Lymph Nodes/*metabolism/pathology
Lymphatic Metastasis
Male
Middle Aged
Neoplasm Recurrence, Local/genetics/metabolism
Receptor, Epidermal Growth Factor/*analysis/genetics
Receptor, erbB-2/*analysis/genetics
Survival Analysis
Figure
Reference
-
1. Ross JS, Fletcher JA. The HER-2/neu oncogene in breast cancer: prognostic factor, predictive factor, and target for therapy. Stem Cells. 1998. 16:413–428.2. Agrawal A, Gutteridge E, Gee JM, Nicholson RI, Robertson JF. Overview of tyrosine kinase inhibitors in clinical breast cancer. Endocr Relat Cancer. 2005. 12:Suppl 1. S135–S144.
Article3. Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001. 411:1017–1021.
Article4. Cobleigh MA, Vogel CL, Tripathy D, Robert NJ, Scholl S, Fehrenbacher L, Wolter JM, Paton V, Shak S, Lieberman G, Slamon DJ. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol. 1999. 17:2639–2648.
Article5. Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, Baselga J, Norton L. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001. 344:783–792.
Article6. Thor A. Are patterns of HER-2/neu amplification and expression among primary tumors and regional metastases indicative of those in distant metastases and predictive of Herceptin response? J Natl Cancer Inst. 2001. 93:1120–1121.
Article7. Chung KY, Shia J, Kemeny NE, Shah M, Schwartz GK, Tse A, Hamilton A, Pan D, Schrag D, Schwartz L, Klimstra DS, Fridman D, Kelsen DP, Saltz LB. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol. 2005. 23:1803–1810.
Article8. Davidoff AM, Kerns BJ, Iglehart JD, Marks JR. Maintenance of p53 alterations throughout breast cancer progression. Cancer Res. 1991. 51:2605–2610.9. Iglehart JD, Kerns BJ, Huper G, Marks JR. Maintenance of DNA content and erbB-2 alterations in intraductal and invasive phases of mammary cancer. Breast Cancer Res Treat. 1995. 34:253–263.
Article10. Foster RS Jr. The biologic and clinical significance of lymphatic metastases in breast cancer. Surg Oncol Clin N Am. 1996. 5:79–104.
Article11. Cardoso F, Di Leo A, Larsimont D, Gancberg D, Rouas G, Dolci S, Ferreira F, Paesmans M, Piccart M. Evaluation of HER2, p53, bcl-2, topoisomerase II-alpha, heat shock proteins 27 and 70 in primary breast cancer and metastatic ipsilateral axillary lymph nodes. Ann Oncol. 2001. 12:615–620.12. Carlsson J, Nordgren H, Sjostrom J, Wester K, Villman K, Bengtsson NO, Ostenstad B, Lundqvist H, Blomqvist C. HER2 expression in breast cancer primary tumours and corresponding metastases. Original data and literature review. Br J Cancer. 2004. 90:2344–2348.
Article13. Gancberg D, Di Leo A, Cardoso F, Rouas G, Pedrocchi M, Paesmans M, Verhest A, Bernard-Marty C, Piccart MJ, Larsimont D. Comparison of HER-2 status between primary breast cancer and corresponding distant metastatic sites. Ann Oncol. 2002. 13:1036–1043.
Article14. Gong Y, Booser DJ, Sneige N. Comparison of HER-2 status determined by fluorescence in situ hybridization in primary and metastatic breast carcinoma. Cancer. 2005. 103:1763–1769.15. Masood S, Bui MM. Assessment of Her-2/neu overexpression in primary breast cancers and their metastatic lesions: an immunohistochemical study. Ann Clin Lab Sci. 2000. 30:259–265.16. Simon R, Nocito A, Hübscher T, Bucher C, Torhorst J, Schraml P, Bubendorf L, Mihatsch MM, Moch H, Wilber K, Schotzau A, Kononen J, Sauter G. Patterns of her-2/neu amplification and overexpression in primary and metastatic breast cancer. J Natl Cancer Inst. 2001. 93:1141–1146.
Article17. Tanner M, Jarvinen P, Isola J. Amplification of HER-2/neu and topoisomerase IIalpha in primary and metastatic breast cancer. Cancer Res. 2001. 61:5345–5348.18. Gupta D, Middleton LP, Whitaker MJ, Abrams J. Comparison of fluorescence and chromogenic in situ hybridization for detection of HER-2/neu oncogene in breast cancer. Am J Clin Pathol. 2003. 119:381–387.
Article19. Allred DC, Harvey JM, Berardo M, Clark GM. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998. 11:155–168.20. Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977. 33:159–174.
Article21. Fidler IJ, Hart IR. Biological diversity in metastatic neoplasms: origins and implications. Science. 1982. 217:998–1003.
Article22. Edgerton SM, Moore D 2nd, Merkel D, Thor AD. erbB-2 (HER-2) and breast cancer progression. Appl Immunohistochem Mol Morphol. 2003. 11:214–221.
Article23. Hao X, Sun B, Hu L, Lahdesmaki H, Dunmire V, Feng Y, Zhang SW, Wang H, Wu C, Wang H, Fuller GN, Symmans WF, Shmulevich I, Zhang W. Differential gene and protein expression in primary breast malignancies and their lymph node metastases as revealed by combined cDNA microarray and tissue microarray analysis. Cancer. 2004. 100:1110–1122.
Article24. Lacroix H, Iglehart JD, Skinner MA, Kraus MH. Overexpression of erbB-2 or EGF receptor proteins present in early stage mammary carcinoma is detected simultaneously in matched primary tumors and regional metastases. Oncogene. 1989. 4:145–151.25. Tsutsui S, Ohno S, Murakami S, Kataoka A, Kinoshita J, Hachitanda Y. EGFR, c-erbB2 and p53 protein in the primary lesions and paired metastatic regional lymph nodes in breast cancer. Eur J Surg Oncol. 2002. 28:383–387.
Article26. Italiano A, Vandenbos FB, Otto J, Mouroux J, Fontaine D, Marcy PY, Cardot N, Thyss A, Pedeutour F. Comparison of the epidermal growth factor receptor gene and protein in primary non-small-cell-lung cancer and metastatic sites: implications for treatment with EGFR-inhibitors. Ann Oncol. 2006. 17:981–985.
Article27. Bhargava R, Gerald WL, Li AR, Pan Q, Lal P, Ladanyi M, Chen B. EGFR gene amplification in breast cancer: correlation with epidermal growth factor receptor mRNA and protein expression and HER-2 status and absence of EGFR-activating mutations. Mod Pathol. 2005. 18:1027–1033.
Article28. Arnold A, Papanikolaou A. Cyclin D1 in breast cancer pathogenesis. J Clin Oncol. 2005. 23:4215–4224.
Article29. Roy PG, Thompson AM. Cyclin D1 and breast cancer. Breast. 2006. 15:718–727.
Article30. Zhu XL, Hartwick W, Rohan T, Kandel R. Cyclin D1 gene amplification and protein expression in benign breast disease and breast carcinoma. Mod Pathol. 1998. 11:1082–1088.31. Naidu R, Wahab NA, Yadav MM, Kutty MK. Expression and amplification of cyclin D1 in primary breast carcinomas: relationship with histopathological types and clinico-pathological parameters. Oncol Rep. 2002. 9:409–416.
Article32. Gillett C, Fantl V, Smith R, Fisher C, Bartek J, Dickson C, Barnes D, Peters G. Amplification and overexpression of cyclin D1 in breast cancer detected by immunohistochemical staining. Cancer Res. 1994. 54:1812–1817.33. Hwang TS, Han HS, Hong YC, Lee HJ, Paik NS. Prognostic value of combined analysis of cyclin D1 and estrogen receptor status in breast cancer patients. Pathol Int. 2003. 53:74–80.
Article34. Jirstrom K, Stendahl M, Ryden L, Kronblad A, Bendahl PO, Stal O, Landberg G. Adverse effect of adjuvant tamoxifen in premenopausal breast cancer with cyclin D1 gene amplification. Cancer Res. 2005. 65:8009–8016.
Article35. Bhargava R, Lal P, Chen B. Feasibility of using tissue microarrays for the assessment of HER-2 gene amplification by fluorescence in situ hybridization in breast carcinoma. Diagn Mol Pathol. 2004. 13:213–216.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Silver-Enhanced In Situ Hybridization as an Alternative to Fluorescence In Situ Hybridization for Assaying HER2 Amplification in Clinical Breast Cancer
- Prognostic Significance COX-2, VEGF and Cyclin D1 in Distant Metastasis of Breast Cancer
- Study of the Relationships between Cyclin D1 and Known Prognostic Factors in Breast Cancer
- Expression of Cyclin D1 and bcl-2 in Infiltrative Ductal Carcinoma of the Breast: Their Correlations and Clinical implications
- Significance on the Expression of Cyclin, MIB-1 in Benign and Malignant Papillary Lesion of the Breast