Korean J Lab Med.
2009 Jun;29(3):231-237. 10.3343/kjlm.2009.29.3.231.
Rapid ABO Genotyping Using Whole Blood without DNA Purification
- Affiliations
-
- 1R&D Center, Lumieye Genetics Co., Ltd., Seoul, Korea.
- 2Department of Laboratory Medicine, Chosun University College of Medicine, Gwangju, Korea.
- 3BioQuest, Inc., Seoul, Korea.
- 4Department of Clinical Laboratory Science, College of Health Sciences, Korea University, Seoul, Korea. swkimkorea@korea.ac.kr
- KMID: 1096975
- DOI: http://doi.org/10.3343/kjlm.2009.29.3.231
Abstract
-
BACKGROUND: ABO genotyping is commonly used in cases of an ABO discrepancy between cell typing and serum typing, as well as in forensic practice for personal identification and paternity testing. We evaluated ABO genotyping via multiplex allele-specific PCR (ASPCR) amplification using whole blood samples without DNA purification.
METHODS
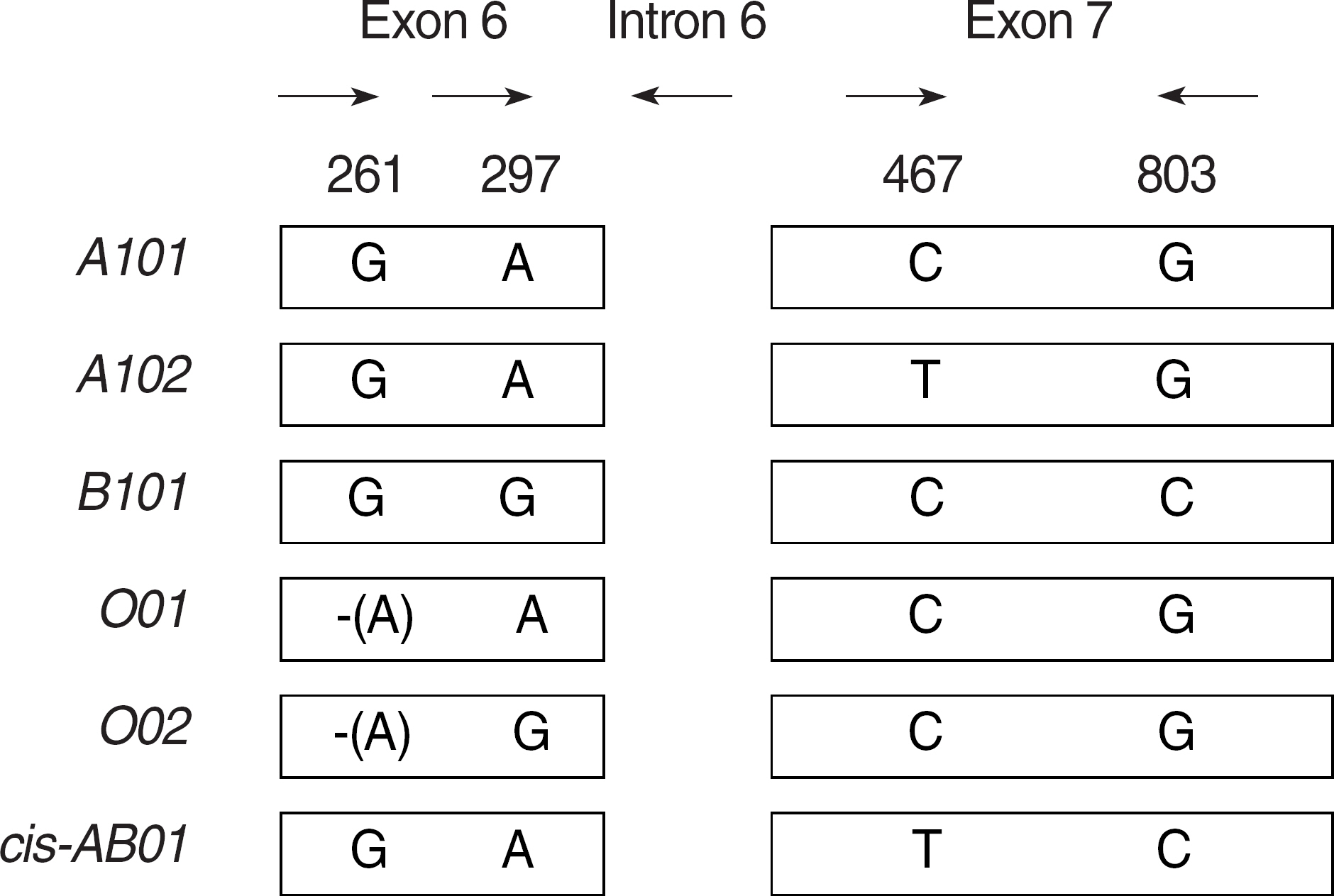
A four-reaction multiplex ASPCR genotyping assay was designed to detect specific nucleotide sequence differences between the six ABO alleles A101, A102, B101, O01, O02, and cis-AB01. The ABO genotypes of 127 randomly chosen samples were determined using the new multiplex ASPCR method.
RESULTS
The genotypes of the 127 samples were found to be A101/A102 (n=1), A102/A102 (n=9), A101/O01 (n=3), A102/O01 (n=12), A102/O02 (n=14), B101/B101 (n=5), B101/O01 (n=18), B101/O02 (n=15), O01/O01 (n=14), O02/O02 (n=8), O01/O02 (n=14) and A102/B101 (n=14), from which phenotypes were calculated to be A (n=39), B (n=38), O (n=36) and AB (n=14). The multiplex ASPCR assay results were compared with the serologically determined blood group phenotypes and genotypes determined by DNA sequencing, and there were no discrepancies.
CONCLUSIONS
This convenient multiplex ASPCR assay, performed using whole blood samples, provides a supplement to routine serological ABO typing and might also be useful in other genotyping applications.
MeSH Terms
Figure
Reference
-
1.Yamamoto F., Clausen H., White T., Marken J., Hakomori S. Molecular genetic basis of the histo-blood group ABO system. Nature. 1990. 345:229–33.
Article2.Yamamoto F., Marken J., Tsuji T., White T., Clausen H., Hakomori S. Cloning and characterization of DNA complementary to human UDP-GalNAc: Fuc alpha 1→2Gal alpha 1→3GalNAc transferase (histo-blood group A transferase) mRNA. J Biol Chem. 1990. 265:1146–51.
Article3.Chang JG., Lee LS., Chen PH., Liu TC., Lin SF., Lee JC. Rapid genotyping of ABO blood group. Blood. 1992. 79:2176–7.4.Lee JC., Chang JG. ABO genotyping by polymerase chain reaction. J Forensic Sci. 1992. 37:1269–75.
Article5.Johnson PH., Hopkinson DA. Detection of ABO blood group polymorphism by denaturing gradient gel electrophoresis. Hum Mol Genet. 1992. 1:341–4.
Article6.Akane A., Yoshimura S., Yoshida M., Okii Y., Watabiki T., Matsubara K, et al. ABO genotyping following a single PCR amplification. J Forensic Sci. 1996. 41:272–4.
Article7.Gassner C., Schmarda A., Nussbaumer W., Schonitzer D. ABO glycosyltransferase genotyping by polymerase chain reaction using sequence-specific primers. Blood. 1996. 88:1852–6.
Article8.Cho D., Jeon MJ., Oh BJ., Song JW., Shin MG., Shin JH, et al. A simplified ABO genotyping by allele-specific polymerase chain reaction. Korean J Lab Med. 2005. 25:123–8. (조덕, 전미정, 오봉준, 송정원, 신명 근, 신종희 등. 대립유전자특이중합효소연쇄반응법에 의한 간편한ABO유전형 검사. 대한진단검사의학회지 2005;25:123-8.).9.Li CX., Pan Q., Guo YG., Li Y., Gao HF., Zhang D, et al. Construction of a multiplex allele-specific PCR-based universal array (ASPUA) and its application to hearing loss screening. Hum Mutat. 2008. 29:306–14.
Article10.Sanchaisuriya K., Chunpanich S., Fucharoen G., Fucharoen S. Multiplex allele-specific PCR assay for differential diagnosis of Hb S, Hb D-Punjab and Hb Tak. Clin Chim Acta. 2004. 343:129–34.
Article11.Angelini A., Di Febbo C., Baccante G., Di Nisio M., Di Ilio C., Cuccurullo F, et al. Identification of three genetic risk factors for venous thrombosis using a multiplex allele-specific PCR assay: comparison of conventional and new alternative methods for the preparation of DNA from clinical samples. J Thromb Thrombolysis. 2003. 16:189–93.
Article12.Yip SP. Single-tube multiplex PCR-SSCP analysis distinguishes 7 common ABO alleles and readily identifies new alleles. Blood. 2000. 95:1487–92.
Article13.Muddiman DC., Null AP., Hannis JC. Precise mass measurement of a double-stranded 500 base-pair (309 kDa) polymerase chain reaction product by negative ion electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun Mass Spectrom. 1999. 13:1201–4.
Article14.Ye S., Dhillon S., Ke X., Collins AR., Day IN. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res. 2001. 29:E88–8.
Article15.Song SH., Chang HE., Ryu KC., Lee HJ., Park KU., Song J, et al. Analysis for eight ABO alleles in Korean population. Korean J Lab Med. 2006. 26:374–9. (송상훈, 장호은, 유광철, 이현정, 박경운, 송정한 등. 한 국인에서 8가지ABO대립유전자의 빈도 분석. 대한진단검사의학회지 2006;26:374-9.).
Article16.Cho D., Kim SH., Jeon MJ., Choi KL., Kee SJ., Shin MG, et al. The serological and genetic basis of the cis-AB blood group in Korea. Vox Sang. 2004. 87:41–3.
Article17.Yang YG., Kim JY., Song YH., Kim DS. A novel buffer system, Any-Direct, can improve polymerase chain reaction from whole blood without DNA isolation. Clin Chim Acta. 2007. 380:112–7.
Article18.Yang YG., Kim JY., Soh MS., Kim DS. A simple and rapid gene amplification from Arabidopsis leaves using AnyDirect system. J Biochem Mol Biol. 2007. 40:444–7.
Article19.Yang YG., Song MK., Park SJ., Kim SW. Direct detection of Shigella flexneri and Salmonella typhimurium in human feces by real-time PCR. J Microbiol Biotechnol. 2007. 17:1616–21.20.Chou Q., Russell M., Birch DE., Raymond J., Bloch W. Prevention of pre-PCR mis-priming and primer dimerization improves low-copy-number amplifications. Nucleic Acids Res. 1992. 11:1717–23.
Article21.Mercier B., Gaucher C., Feugeas O., Mazurier C. Direct PCR from whole blood, without DNA extraction. Nucleic Acids Res. 1990. 18:5908.
Article22.Ohhara M., Kurosu Y., Esumi M. Direct PCR of whole blood and hair shafts by microwave treatment. Biotechniques. 1994. 17:726–8.23.Panaccio M., Georgesz M., Lew AM. FoLT PCR: a simple PCR protocol for amplifying DNA directly from whole blood. Biotechniques. 1993. 14:238–43.24.Nishimura N., Nakayama T., Tonoike H., Kojima K., Kato S. Direct polymerase chain reaction from whole blood without DNA isolation. Ann Clin Biochem. 2000. 37:674–80.