Korean J Lab Med.
2010 Apr;30(2):190-194. 10.3343/kjlm.2010.30.2.190.
Molecular Characterization of the NF2 Gene in Korean Patients with Neurofibromatosis Type 2: A Report of Four Novel Mutations
- Affiliations
-
- 1Department of Laboratory Medicine, Seoul National University Hospital, Seoul, Korea. sparkle@snu.ac.kr
- 2Department of Neurosurgery, Seoul National University Hospital, Seoul, Korea.
- 3Clinical Research Institute, Seoul National University Hospital, Seoul, Korea.
- 4Department of Laboratory Medicine, National Cancer Center, Goyang, Korea.
- KMID: 1096807
- DOI: http://doi.org/10.3343/kjlm.2010.30.2.190
Abstract
- BACKGROUND
Neurofibromatosis type 2 (NF2) is an autosomal dominant syndrome caused by the NF2 tumor suppressor gene. However, the NF2 mutation characteristics in Korean patients are not sufficiently understood. In this study, we conducted a comprehensive mutational analysis in 7 Korean NF2 patients by performing direct sequencing and gene-dosage assessment.
METHODS
We analyzed all exons and flanking regions of NF2 by direct sequencing and screened the deletions or duplications involving NF2 by multiplex ligation-dependent probe amplification.
RESULTS
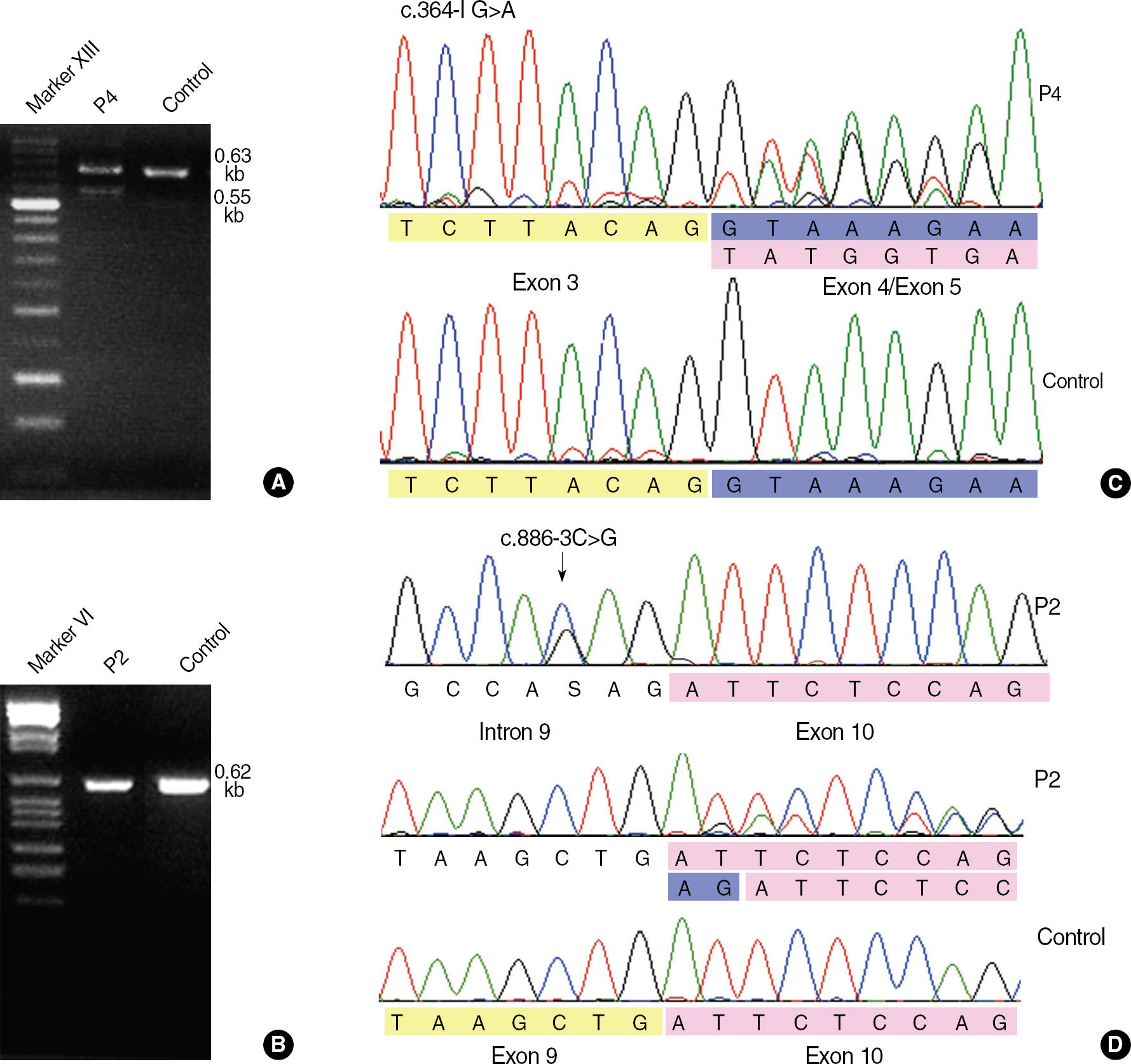
Four novel NF2 mutations, including 2 splice-site mutations (c.364-1G>A and c.886-3C>G), 1 frameshift mutation (c.524delA), and 1 missense mutation (c.397T>C; p.Cys133Arg), were identified in our patients. No large deletion or duplication was identified in our series. Subsequently, we identified an abnormal splicing product by using reverse transcription-PCR and direct sequencing in 2 patients with a novel splice-site mutation. The missense mutation c.397T>C was predicted to have harmful effects on protein function.
CONCLUSIONS
The detection rate of NF2 mutations in Korean patients (57%) is similar to those in other populations. Our results provided a greater insight into the mutational spectrum of the NF2 gene in Korean subjects.
Keyword
MeSH Terms
-
3' Flanking Region/genetics
5' Flanking Region/genetics
Adult
Aged
Amino Acid Sequence
Asian Continental Ancestry Group/*genetics
Child, Preschool
Exons
Female
Frameshift Mutation
*Genes, Neurofibromatosis 2
Humans
Male
Middle Aged
Molecular Sequence Data
*Mutation
Mutation, Missense
Neurofibromatosis 2/diagnosis/*genetics
RNA Splice Sites
Republic of Korea
Sequence Analysis, DNA
Young Adult
Figure
Reference
-
1.Evans DG., Huson SM., Donnai D., Neary W., Blair V., Newton V, et al. A clinical study of type 2 neurofibromatosis. Q J Med. 1992. 84:603–18.2.Wallace AJ., Watson CJ., Oward E., Evans DG., Elles RG. Mutation scanning of the NF2 gene: an improved service based on meta-PCR/sequencing, dosage analysis, and loss of heterozygosity analysis. Genet Test. 2004. 8:368–80.3.Kluwe L., Nygren AO., Errami A., Heinrich B., Matthies C., Tatagiba M, et al. Screening for large mutations of the NF2 gene. Genes Chromosomes Cancer. 2005. 42:384–91.4.Yang HJ., Won YJ., Park KJ., Jung HW., Choi KS., Park JG. Germline mutations of the NF2 gene in Korean neurofibromatosis 2 patient. J Korean Cancer Assoc. 1998. 30:790–9.5.Ferrer-Costa C., Orozco M., de la Cruz X. Sequence-based prediction of pathological mutations. Proteins. 2004. 57:811–9.
Article6.Ng PC., Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001. 11:863–74.
Article7.Ramensky V., Bork P., Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002. 30:3894–900.
Article8.Ahronowitz I., Xin W., Kiely R., Sims K., MacCollin M., Nunes FP. Mutational spectrum of the NF2 gene: a meta-analysis of 12 years of research and diagnostic laboratory findings. Hum Mutat. 2007. 28:1–12.9.Evans DG., Trueman L., Wallace A., Collins S., Strachan T. Genotype/phenotype correlations in type 2 neurofibromatosis (NF2): evidence for more severe disease associated with truncating mutations. J Med Genet. 1998. 35:450–5.
Article10.Baser ME., Kuramoto L., Joe H., Friedman JM., Wallace AJ., Gillespie JE, et al. Genotype-phenotype correlations for nervous system tumors in neurofibromatosis 2: a population-based study. Am J Hum Genet. 2004. 75:231–9.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Germline Mutations of the NF2 Gene in Korean Neurofibromatosis 2 Patient
- General anesthesia for emergency cesarean section in a parturient with neurofibromatosis type 2: A case report
- Merlin; NF2 Tumor Suppressor and Regulator of Receptor Distribution/Signaling
- Genetic and clinical characteristics of Korean patients with neurofibromatosis type 2
- Analysis of Genetic Mutations and the Clinical Manifestations of Neurofibromatosis Type 2 in Korea: A Single Institutional Experience