Cancer Res Treat.
2025 Apr;57(2):597-611. 10.4143/crt.2024.675.
Clinical Impact of Microbiome Characteristics in Treatment-Naïve Extranodal NK/T-Cell Lymphoma Patients
- Affiliations
-
- 1Division of Hematology-Oncology, Department of Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- 2CJ Bioscience Inc., Seoul, Korea
- 3Department of Pathology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- KMID: 2566876
- DOI: http://doi.org/10.4143/crt.2024.675
Abstract
- Purpose
Extranodal natural killer/T-cell lymphoma (ENKTL) predominantly manifests in East Asia and Latin America. Despite shared intrinsic factors, such as ethnic and genetic backgrounds, the progression of ENKTL can be influenced by extrinsic factors related to changing lifestyle patterns.
Materials and Methods
This study collected stool samples from newly diagnosed (ND)–ENKTL patients (n=40) and conducted whole genome shotgun sequencing.
Results
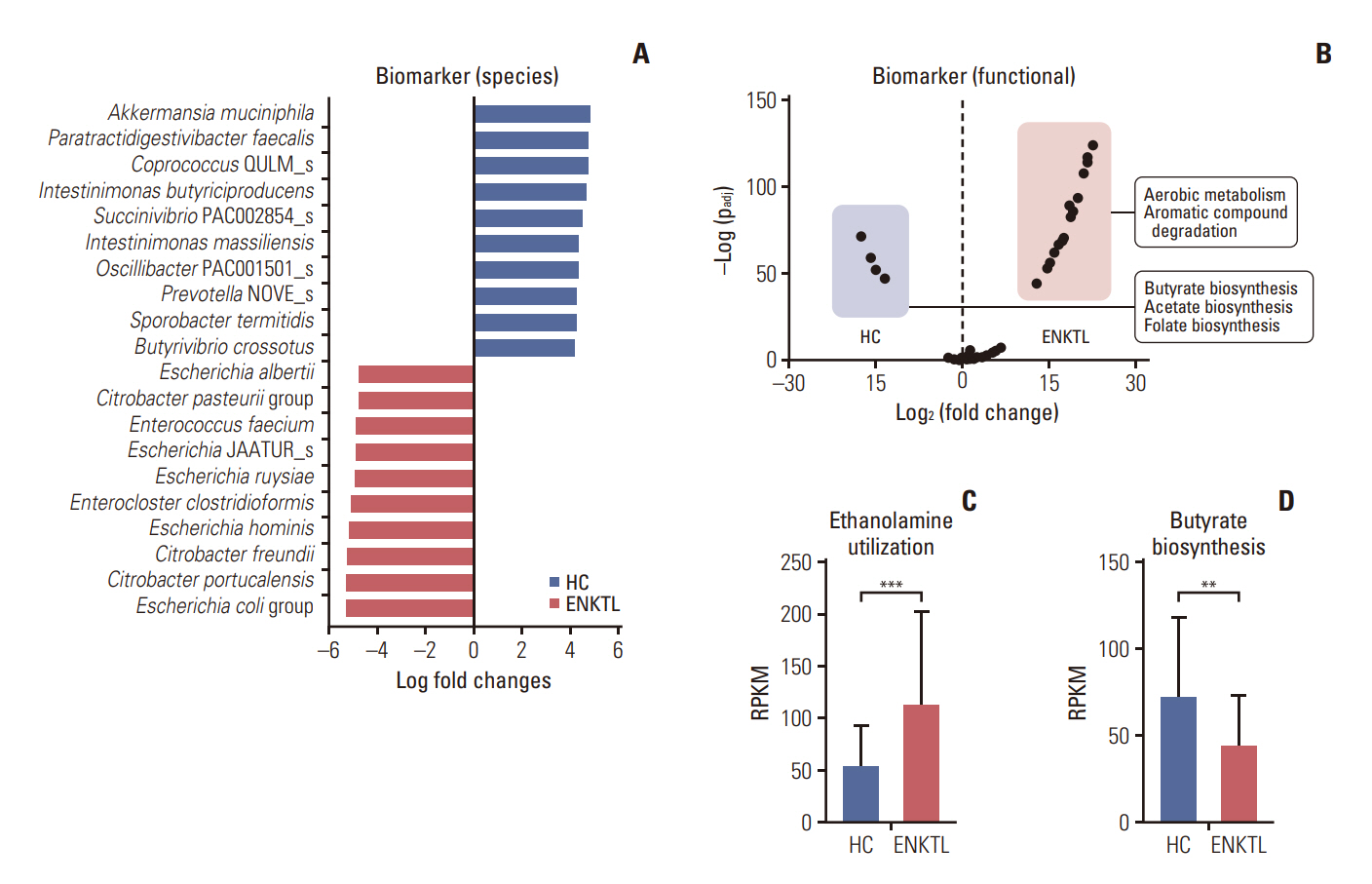
ND-ENKTL revealed reduced alpha diversity in ND-ENKTL compared to healthy controls (HCs) (p=0.008), with Enterobacteriaceae abundance significantly contributing to the beta diversity difference between ENKTL and HCs (p < 0.001). Functional analysis indicated upregulated aerobic metabolism and degradation of aromatic compounds in ND-ENKTL. Enterobacteriaceae were associated not only with clinical data explaining disease status (serum C-reactive protein, stage, prognosis index of natural killer cell lymphoma [PINK], and PINK-E) but also with clinical outcomes (early relapse and short progression-free survival). The relative abundance of Enterobacteriaceae at the family level was similar between ENKTL and diffuse large B-cell lymphoma (DLBCL) (p=0.140). However, the ENKTL exhibited a higher abundance of Escherichia, in contrast to the prevalence of Enterobacter and Citrobacter in DLBCL. Linear regression analysis demonstrated a significant association between Escherichia abundance and programmed cell death-ligand-1 (PD-L1) levels in tissue samples (p=0.025), whereas no correlation with PD-L1 was observed for Enterobacteriaceae at the family level (p=0.571).
Conclusion
ND-ENKTL exhibited an abundance of Enterobacteriaceae and a dominant presence of Escherichia. These microbial characteristics correlated with disease status, treatment outcomes, and PD-L1 expression, suggesting the potential of the ENKTL microbiome as a biomarker and cause of lymphomagenesis, which warrants further exploration.
Keyword
Figure
Reference
-
References
1. Damania B, Kenney SC, Raab-Traub N. Epstein-Barr virus: biology and clinical disease. Cell. 2022; 185:3652–70.
Article2. Tse E, Fox CP, Glover A, Yoon SE, Kim WS, Kwong YL. Extranodal natural killer/T-cell lymphoma: an overview on pathology and clinical management. Semin Hematol. 2022; 59:198–209.
Article3. Lin GW, Xu C, Chen K, Huang HQ, Chen J, Song B, et al. Genetic risk of extranodal natural killer T-cell lymphoma: a genome-wide association study in multiple populations. Lancet Oncol. 2020; 21:306–16.4. Li Z, Xia Y, Feng LN, Chen JR, Li HM, Cui J, et al. Genetic risk of extranodal natural killer T-cell lymphoma: a genome-wide association study. Lancet Oncol. 2016; 17:1240–7.
Article5. Vose J, Armitage J, Weisenburger D; International T-Cell Lymphoma Project. International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol. 2008; 26:4124–30.
Article6. Yoon SE, Song Y, Kim SJ, Yoon DH, Chen TY, Koh Y, et al. Comprehensive analysis of peripheral T-cell and natural killer/T-cell lymphoma in Asian patients: a multinational, multicenter, prospective registry study in Asia. Lancet Reg Health West Pac. 2021; 10:100126.
Article7. Gupta VK, Paul S, Dutta C. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front Microbiol. 2017; 8:1162.
Article8. Shi Z, Li X, Wang X, Zhang L, Li L, Fu X, et al. Characteristics and clinical implications of the nasal microbiota in extranodal NK/T-cell lymphoma, nasal type. Front Cell Infect Microbiol. 2021; 11:686595.
Article9. Shi Z, Hu G, Li MW, Zhang L, Li X, Li L, et al. Gut microbiota as non-invasive diagnostic and prognostic biomarkers for natural killer/T-cell lymphoma. Gut. 2023; 72:1999–2002.
Article10. Yoon SE, Kang W, Choi S, Park Y, Chalita M, Kim H, et al. The influence of microbial dysbiosis on immunochemotherapy-related efficacy and safety in diffuse large B-cell lymphoma. Blood. 2023; 141:2224–38.
Article11. Kim SJ, Yoon DH, Jaccard A, Chng WJ, Lim ST, Hong H, et al. A prognostic index for natural killer cell lymphoma after non-anthracycline-based treatment: a multicentre, retrospective analysis. Lancet Oncol. 2016; 17:389–400.
Article12. Cho J, Kim SJ, Park WY, Kim J, Woo J, Kim G, et al. Immune subtyping of extranodal NK/T-cell lymphoma: a new biomarker and an immune shift during disease progression. Mod Pathol. 2020; 33:603–15.
Article13. Cheson BD, Pfistner B, Juweid ME, Gascoyne RD, Specht L, Horning SJ, et al. Revised response criteria for malignant lymphoma. J Clin Oncol. 2007; 25:579–86.
Article14. Sepich-Poore GD, Zitvogel L, Straussman R, Hasty J, Wargo JA, Knight R. The microbiome and human cancer. Science. 2021; 371:eabc4552.
Article15. Park EM, Chelvanambi M, Bhutiani N, Kroemer G, Zitvogel L, Wargo JA. Targeting the gut and tumor microbiota in cancer. Nat Med. 2022; 28:690–703.
Article16. Upadhyay Banskota S, Skupa SA, El-Gamal D, D’Angelo CR. Defining the role of the gut microbiome in the pathogenesis and treatment of lymphoid malignancies. Int J Mol Sci. 2023; 24:2309.
Article17. Hamerman JA, Ogasawara K, Lanier LL. NK cells in innate immunity. Curr Opin Immunol. 2005; 17:29–35.
Article18. Bajor-Dattilo EB, Pittaluga S, Jaffe ES. Pathobiology of T-cell and NK-cell lymphomas. Best Pract Res Clin Haematol. 2013; 26:75–87.
Article19. Kers JG, Saccenti E. The power of microbiome studies: some considerations on which alpha and beta metrics to use and how to report results. Front Microbiol. 2021; 12:796025.
Article20. Helmink BA, Khan MA, Hermann A, Gopalakrishnan V, Wargo JA. The microbiome, cancer, and cancer therapy. Nat Med. 2019; 25:377–88.
Article21. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018; 359:97–103.22. Yuan L, Wang W, Zhang W, Zhang Y, Wei C, Li J, et al. Gut microbiota in untreated diffuse large B cell lymphoma patients. Front Microbiol. 2021; 12:646361.
Article23. Diefenbach CS, Peters BA, Li H, Raphael B, Moskovits T, Hymes K, et al. Microbial dysbiosis is associated with aggressive histology and adverse clinical outcome in B-cell nonHodgkin lymphoma. Blood Adv. 2021; 5:1194–8.
Article24. Rivera-Chavez F, Lopez CA, Baumler AJ. Oxygen as a driver of gut dysbiosis. Free Radic Biol Med. 2017; 105:93–101.
Article25. Litvak Y, Byndloss MX, Baumler AJ. Colonocyte metabolism shapes the gut microbiota. Science. 2018; 362:eaat9076.
Article26. Zeng MY, Inohara N, Nunez G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017; 10:18–26.
Article27. Liu XF, Shao JH, Liao YT, Wang LN, Jia Y, Dong PJ, et al. Reglation of short-chain fatty acids in the immune system. Front Immunol. 2023; 14:1186892.28. Mikkelsen K, Apostolopoulos V. Vitamin B12, folic acid, and the immune system. In : Mahmoudi M, Razaei N, editors. Nutrition and immunity. Springer;2019. p. 103–14.29. Li YJ, Li ZM, Xia Y, Huang JJ, Huang HQ, Xia ZJ, et al. Serum C-reactive protein (CRP) as a simple and independent prognostic factor in extranodal natural killer/T-cell lymphoma, nasal type. PLoS One. 2013; 8:e64158.
Article30. Bao C, Zhou D, Zhu L, Qian W, Ye X. Increased serum level of interleukin-6 correlates with negative prognostic factors in extranodal NK/T-cell lymphoma. Transl Cancer Res. 2020; 9:2378–89.
Article31. Mitsunaga S, Ikeda M, Shimizu S, Ohno I, Takahashi H, Okuyama H, et al. C-reactive protein level is an indicator of the aggressiveness of advanced pancreatic cancer. Pancreas. 2016; 45:110–6.
Article32. Troppan KT, Schlick K, Deutsch A, Melchardt T, Egle A, Stojakovic T, et al. C-reactive protein level is a prognostic indicator for survival and improves the predictive ability of the R-IPI score in diffuse large B-cell lymphoma patients. Br J Cancer. 2014; 111:55–60.
Article33. Song TL, Nairismagi ML, Laurensia Y, Lim JQ, Tan J, Li ZM, et al. Oncogenic activation of the STAT3 pathway drives PD-L1 expression in natural killer/T-cell lymphoma. Blood. 2018; 132:1146–58.34. Li X, Cheng Y, Zhang M, Yan J, Li L, Fu X, et al. Activity of pembrolizumab in relapsed/refractory NK/T-cell lymphoma. J Hematol Oncol. 2018; 11:15.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Recent updates on extranodal NK/T-cell lymphoma
- Extranodal NK/T Cell Lymphoma, Nasal Type that Occurred in Patients with Atrophic Rhinitis
- A Case of Extranodal NK/T-cell Lymphoma at the Base of Tongue
- A Case of Extranodal NK/T Cell Lymphoma, Nasal Type with Cutaneous Involvement
- Extranodal NK/T cell lymphoma