Cancer Res Treat.
2024 Apr;56(2):442-454. 10.4143/crt.2023.764.
Genomic Landscape of Pulmonary Sarcomatoid Carcinoma
- Affiliations
-
- 1Department of Pathology, Seoul National University College of Medicine, Seoul, Korea
- 2Department of Pathology and Translational Medicine, Seoul National University Bundang Hospital, Seongnam, Korea
- 3Artificial Intelligence Institute of Seoul National University, Seoul, Korea
- 4Genomic Medicine Institute, Seoul National University Medical Research Center, Seoul, Korea
- KMID: 2554335
- DOI: http://doi.org/10.4143/crt.2023.764
Abstract
- Purpose
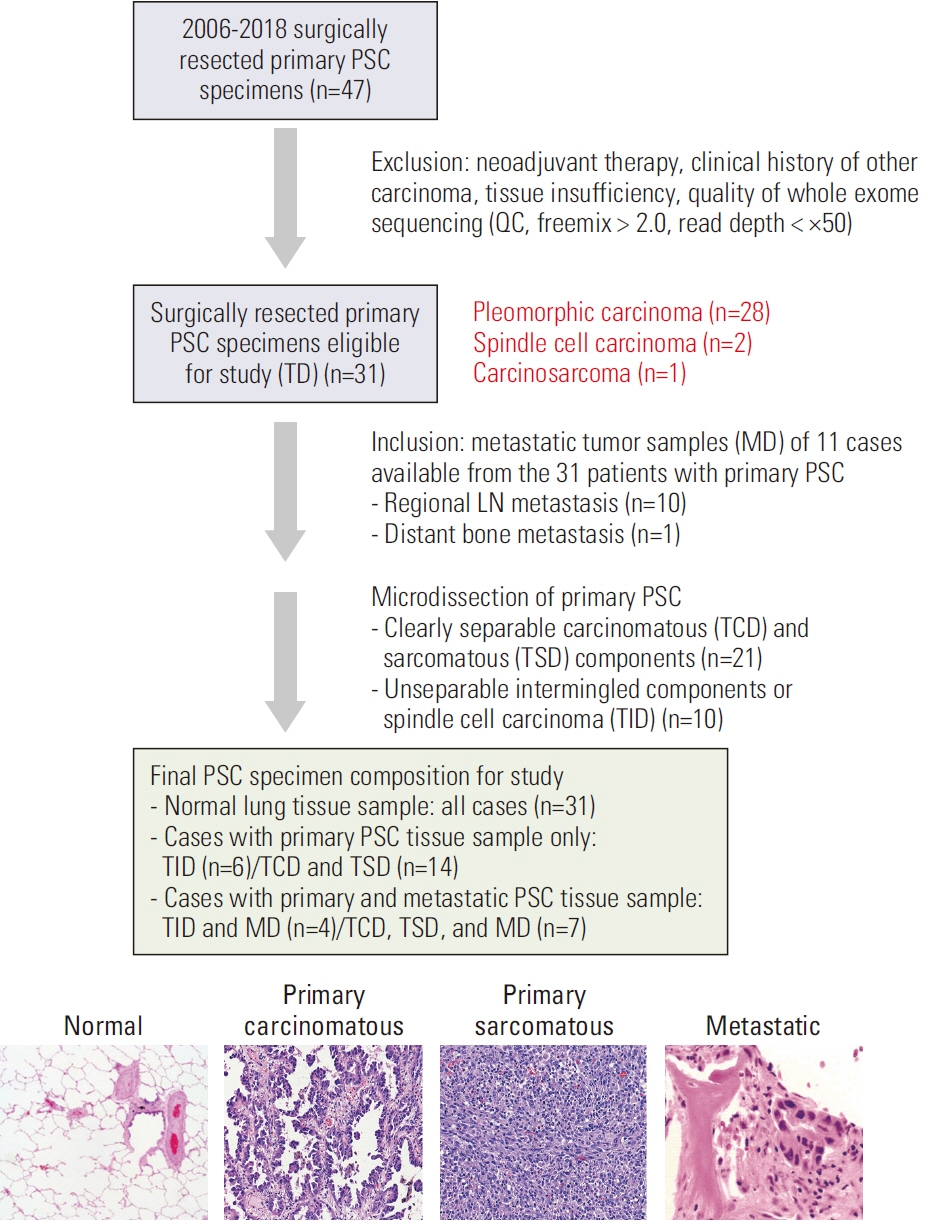
Pulmonary sarcomatoid carcinoma (PSC) is a rare aggressive subtype of non–small cell lung cancer (NSCLC) with limited therapeutic strategies. We attempted to elucidate the evolutionary trajectories of PSC using multiregional and longitudinal tumor samples.
Materials and Methods
A total of 31 patients were enrolled in this study and 11 longitudinal samples were available from them. Using whole exome sequencing data, we analyzed the mutational signatures in both carcinomatous and sarcomatous areas in primary tumors of the 31 patients and longitudinal samples obtained from 11 patients. Furthermore, digital droplet polymerase chain reaction (ddPCR), and programmed death-ligand 1 (PD-L1) immunohistochemistry using the Ventana SP263 assay were performed.
Results
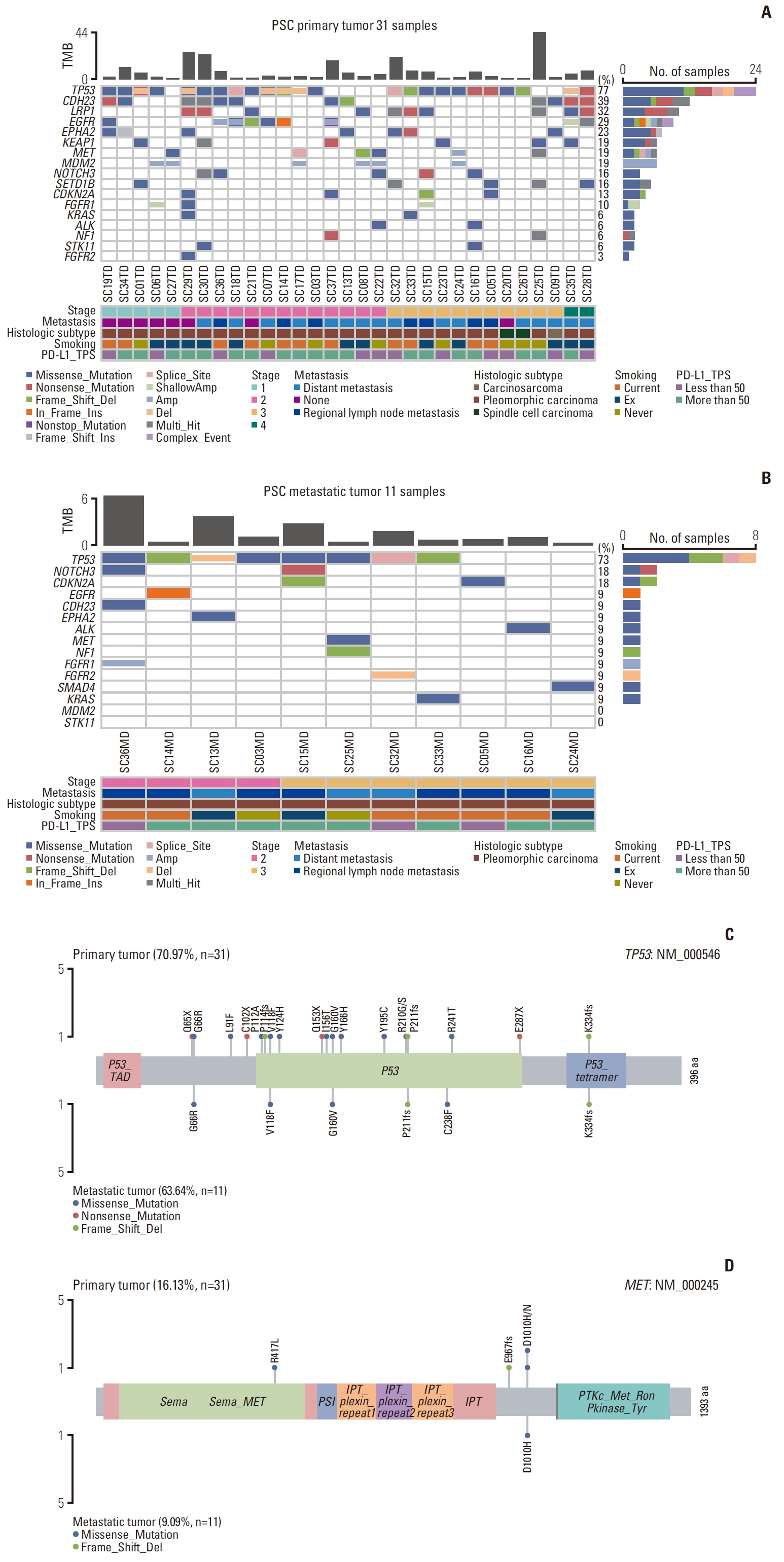
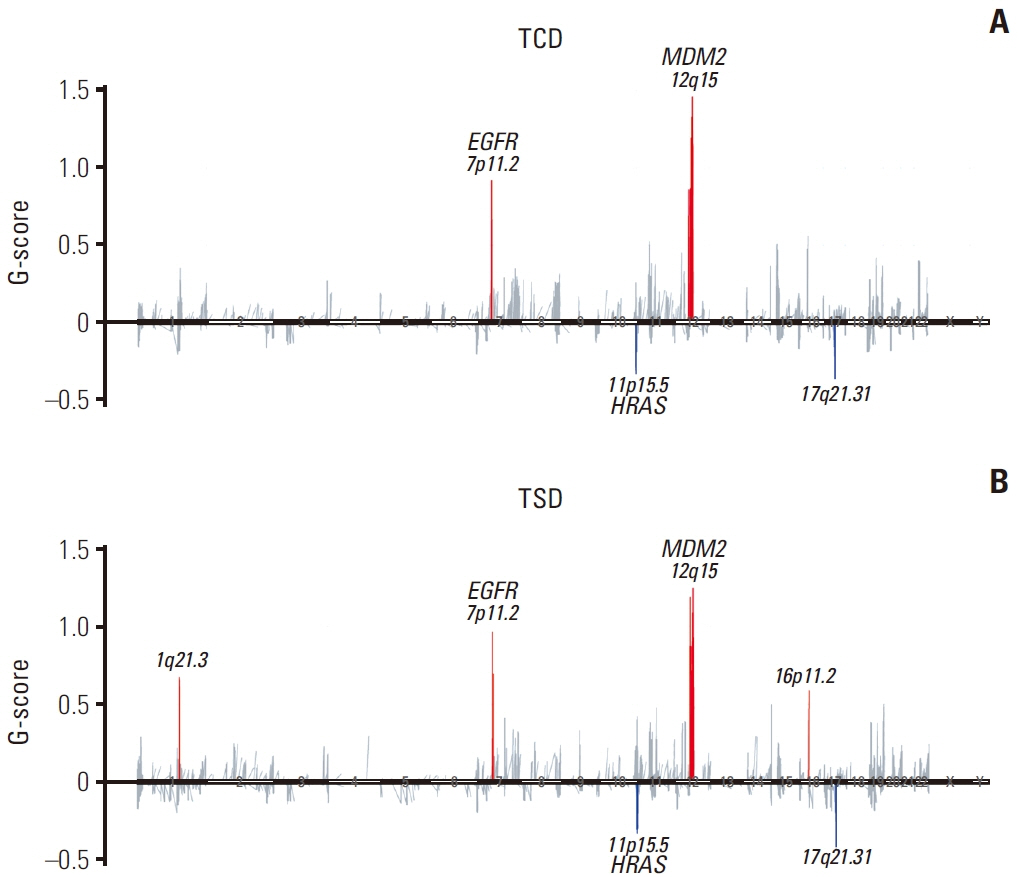
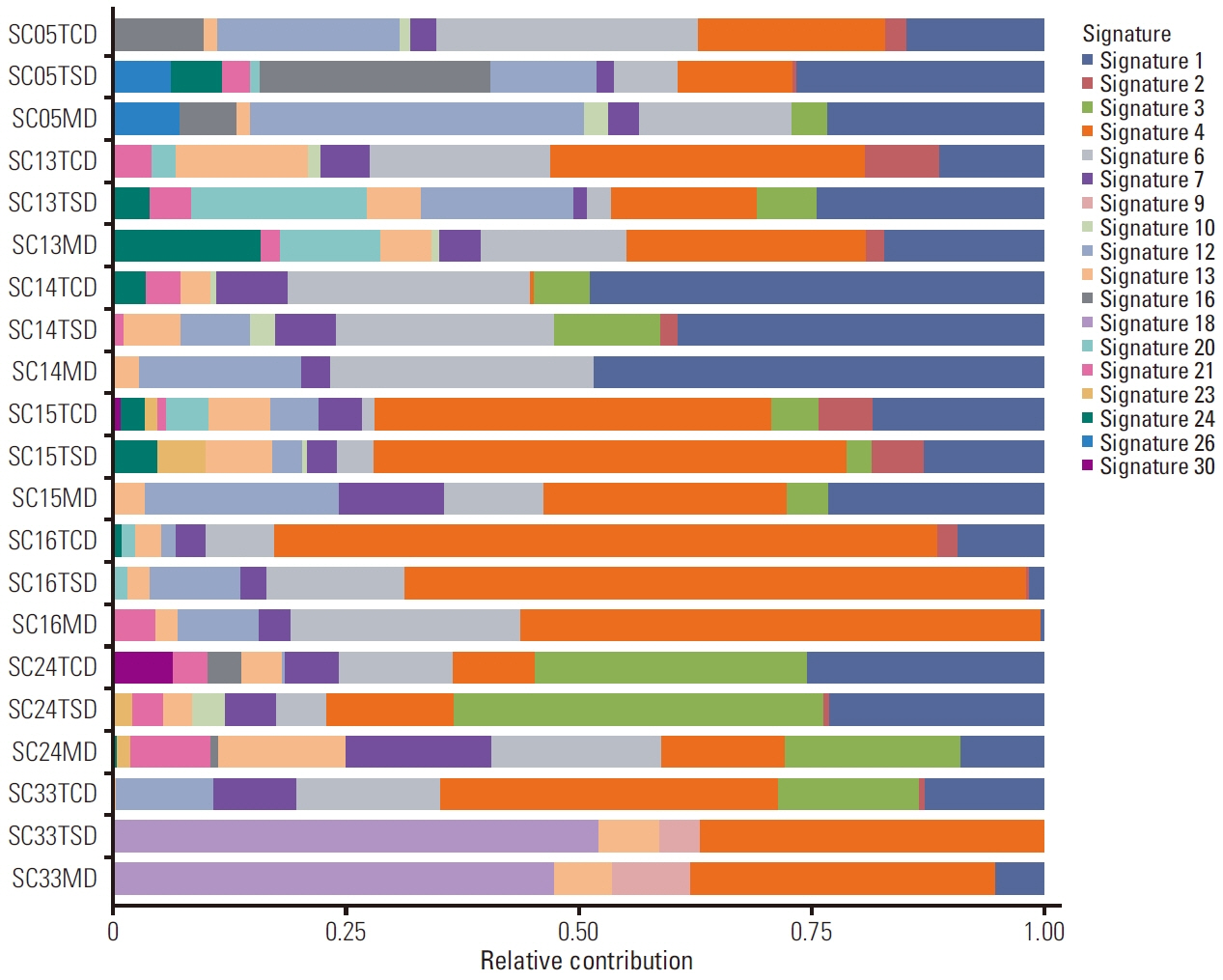
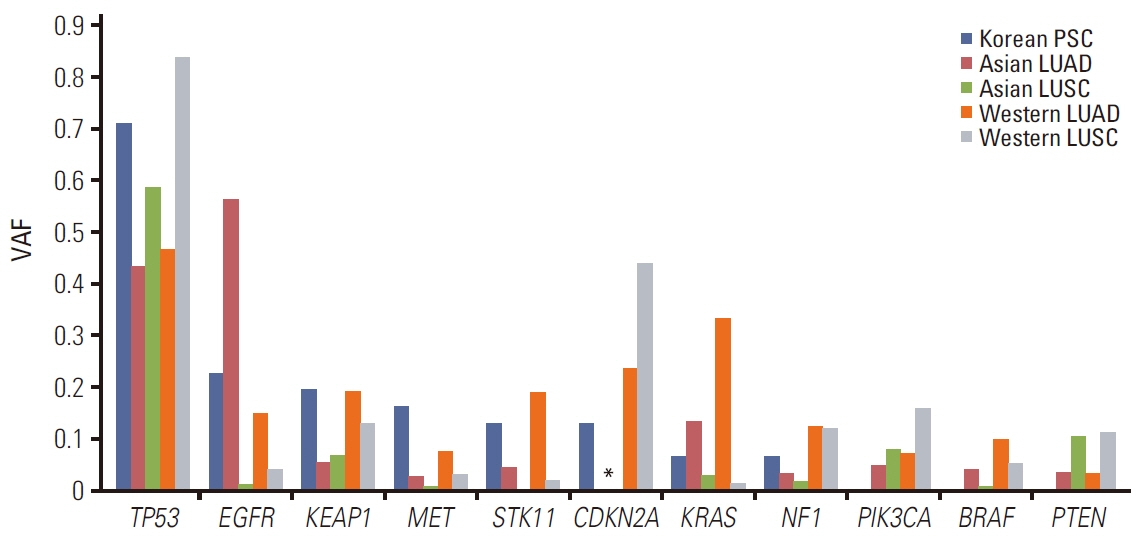
TP53 was identified as the most frequently altered gene in the primary (74%) and metastatic (73%) samples. MET exon 14 skipping mutations, confirmed by ddPCR, and TP53 mutations were mutually exclusive; whereas, MET exon 14 skipping mutations frequently co-occurred with MDM2 amplification. Metastatic tumors showed dissimilar genetic profiles from either primary component. During metastasis, the signatures of APOBEC decreased in metastatic lesions compared with that in primary lesions. PSC showed higher MET and KEAP1 mutations and stronger PD-L1 protein expression compared with that recorded in other NSCLCs.
Conclusion
Decreased APOBEC signatures and subclonal diversity were detected during malignant progression in PSC. Frequent MET mutations and strong PD-L1 expression distinguished PSC from other NSCLCs. The aggressiveness and therapeutic difficulties of PSC were possibly attributable to profound intratumoral and intertumoral genetic diversity. Next-generation sequencing could suggest the appropriate treatment strategy for PSC.
Keyword
Figure
Reference
-
References
1. Travis WD, Brambilla E, Burk AP, Marx A, Nicholson AG. WHO classification of tumours of the lung, pleura, thymus and heart. 4th ed. Lyon: IARC Press;2015.2. Lococo F, Torricelli F, Rossi G, Alifano M, Damotte D, Rapicetta C, et al. Inter-relationship between PD-L1 expression and clinic-pathological features and driver gene mutations in pulmonary sarcomatoid carcinomas. Lung Cancer. 2017; 113:93–101.
Article3. Liang X, Cheng Y, Yuan Z, Yan Z, Li Q, Huang Y, et al. Clinical, pathological and treatment factors associated with the survival of patients with pulmonary sarcomatoid carcinoma. Oncol Lett. 2020; 19:4031–9.
Article4. Vieira T, Girard N, Ung M, Monnet I, Cazes A, Bonnette P, et al. Efficacy of first-line chemotherapy in patients with advanced lung sarcomatoid carcinoma. J Thorac Oncol. 2013; 8:1574–7.
Article5. Yang J, Antin P, Berx G, Blanpain C, Brabletz T, Bronner M, et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2020; 21:341–52.
Article6. Lococo F, Gandolfi G, Rossi G, Pinto C, Rapicetta C, Cavazza A, et al. Deep sequencing analysis reveals that KRAS mutation is a marker of poor prognosis in patients with pulmonary sarcomatoid carcinoma. J Thorac Oncol. 2016; 11:1282–92.
Article7. Terra SB, Jang JS, Bi L, Kipp BR, Jen J, Yi ES, et al. Molecular characterization of pulmonary sarcomatoid carcinoma: analysis of 33 cases. Mod Pathol. 2016; 29:824–31.
Article8. Liu X, Jia Y, Stoopler MB, Shen Y, Cheng H, Chen J, et al. Nextgeneration sequencing of pulmonary sarcomatoid carcinoma reveals high frequency of actionable MET gene mutations. J Clin Oncol. 2016; 34:794–802.
Article9. Yang Z, Xu J, Li L, Li R, Wang Y, Tian Y, et al. Integrated molecular characterization reveals potential therapeutic strategies for pulmonary sarcomatoid carcinoma. Nat Commun. 2020; 11:4878.
Article10. Velcheti V, Rimm DL, Schalper KA. Sarcomatoid lung carcinomas show high levels of programmed death ligand-1 (PDL1). J Thorac Oncol. 2013; 8:803–5.
Article11. Domblides C, Leroy K, Monnet I, Mazieres J, Barlesi F, Gounant V, et al. Efficacy of immune checkpoint inhibitors in lung sarcomatoid carcinoma. J Thorac Oncol. 2020; 15:860–6.
Article12. Amin MB, Edge SB, Greene FL, Byrd DR, Brookland RK, Washington MK, et al. AJCC cancer staging manual. 8th ed. New York: Springer;2017.13. Zhao M, Liu Y, Zheng C, Qu H. dbEMT 2.0: an updated database for epithelial-mesenchymal transition genes with experimentally verified information and precalculated regulation information for cancer metastasis. J Genet Genomics. 2019; 46:595–7.
Article14. Zhao M, Kong L, Liu Y, Qu H. dbEMT: an epithelial-mesenchymal transition associated gene resource. Sci Rep. 2015; 5:11459.
Article15. Favero F, Joshi T, Marquard AM, Birkbak NJ, Krzystanek M, Li Q, et al. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann Oncol. 2015; 26:64–70.
Article16. Yu Y, Chen R, Zhao J, Yi X, Lu S. Analysis of canonical and noncanonical splicing site mutation of MET that causes exon 14 skipping. J Clin Oncol. 2020; 38(15 Suppl):e21513.
Article17. Awad MM, Oxnard GR, Jackman DM, Savukoski DO, Hall D, Shivdasani P, et al. MET exon 14 mutations in non-smallcell lung cancer are associated with advanced age and stagedependent MET genomic amplification and c-Met overexpression. J Clin Oncol. 2016; 34:721–30.18. Chen J, Yang H, Teo AS, Amer LB, Sherbaf FG, Tan CQ, et al. Genomic landscape of lung adenocarcinoma in East Asians. Nat Genet. 2020; 52:177–86.
Article19. Li C, Gao Z, Li F, Li X, Sun Y, Wang M, et al. Whole exome sequencing identifies frequent somatic mutations in cell-cell adhesion genes in Chinese patients with lung squamous cell carcinoma. Sci Rep. 2015; 5:14237.
Article20. Mehrad M, Roy S, LaFramboise WA, Petrosko P, Miller C, Incharoen P, et al. KRAS mutation is predictive of outcome in patients with pulmonary sarcomatoid carcinoma. Histopathology. 2018; 73:207–14.
Article21. Nakagomi T, Goto T, Hirotsu Y, Shikata D, Yokoyama Y, Higuchi R, et al. New therapeutic targets for pulmonary sarcomatoid carcinomas based on their genomic and phylogenetic profiles. Oncotarget. 2018; 9:10635–49.
Article22. Li X, Wu D, Liu H, Chen J. Pulmonary sarcomatoid carcinoma: progress, treatment and expectations. Ther Adv Med Oncol. 2020; 12:1758835920950207.
Article23. Salgia R, Sattler M, Scheele J, Stroh C, Felip E. The promise of selective MET inhibitors in non-small cell lung cancer with MET exon 14 skipping. Cancer Treat Rev. 2020; 87:102022.
Article24. Kim S, Koh J, Kim MY, Kwon D, Go H, Kim YA, et al. PD-L1 expression is associated with epithelial-to-mesenchymal transition in adenocarcinoma of the lung. Hum Pathol. 2016; 58:7–14.
Article25. Dong P, Xiong Y, Yue J, Hanley SJ, Watari H. Tumor-intrinsic PD-L1 signaling in cancer initiation, development and treatment: beyond immune evasion. Front Oncol. 2018; 8:386.
Article26. Goh JY, Feng M, Wang W, Oguz G, Yatim S, Lee PL, et al. Chromosome 1q21.3 amplification is a trackable biomarker and actionable target for breast cancer recurrence. Nat Med. 2017; 23:1319–30.
Article27. Wang S, Jia M, He Z, Liu XS. APOBEC3B and APOBEC mutational signature as potential predictive markers for immunotherapy response in non-small cell lung cancer. Oncogene. 2018; 37:3924–36.
Article28. Chen H, Chong W, Teng C, Yao Y, Wang X, Li X. The immune response-related mutational signatures and driver genes in non-small-cell lung cancer. Cancer Sci. 2019; 110:2348–56.
Article29. Selenica P, Marra A, Choudhury NJ, Gazzo A, Falcon CJ, Patel J, et al. APOBEC mutagenesis, kataegis, chromothripsis in EGFR-mutant osimertinib-resistant lung adenocarcinomas. Ann Oncol. 2022; 33:1284–95.
Article30. Venkatesan S, Rosenthal R, Kanu N, McGranahan N, Bartek J, Quezada SA, et al. Perspective: APOBEC mutagenesis in drug resistance and immune escape in HIV and cancer evolution. Ann Oncol. 2018; 29:563–72.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Sarcomatoid Carcinoma of the Lung: Two cases report
- Sarcomatoid Renal Cell Carcinoma; Special Reference to its Distinction from Carcinosarcoma
- A Case of Sarcomatoid Renal Cell Carcinoma with Multiple Renal Stones and Pyonephrosis
- Sarcomatoid urothelial carcinoma arising in the female urethral diverticulum
- A Case of Sarcomatoid Carcinoma of Larynx