Korean J Physiol Pharmacol.
2024 Mar;28(2):113-120. 10.4196/kjpp.2024.28.2.113.
Transcriptional regulation of genetic variants in the SLC40A1 promoter
- Affiliations
-
- 1Department of Pharmacology, Inflammation-Cancer Microenvironment Research Center, College of Medicine, Ewha Womans University, Seoul 07804, Korea
- KMID: 2553523
- DOI: http://doi.org/10.4196/kjpp.2024.28.2.113
Abstract
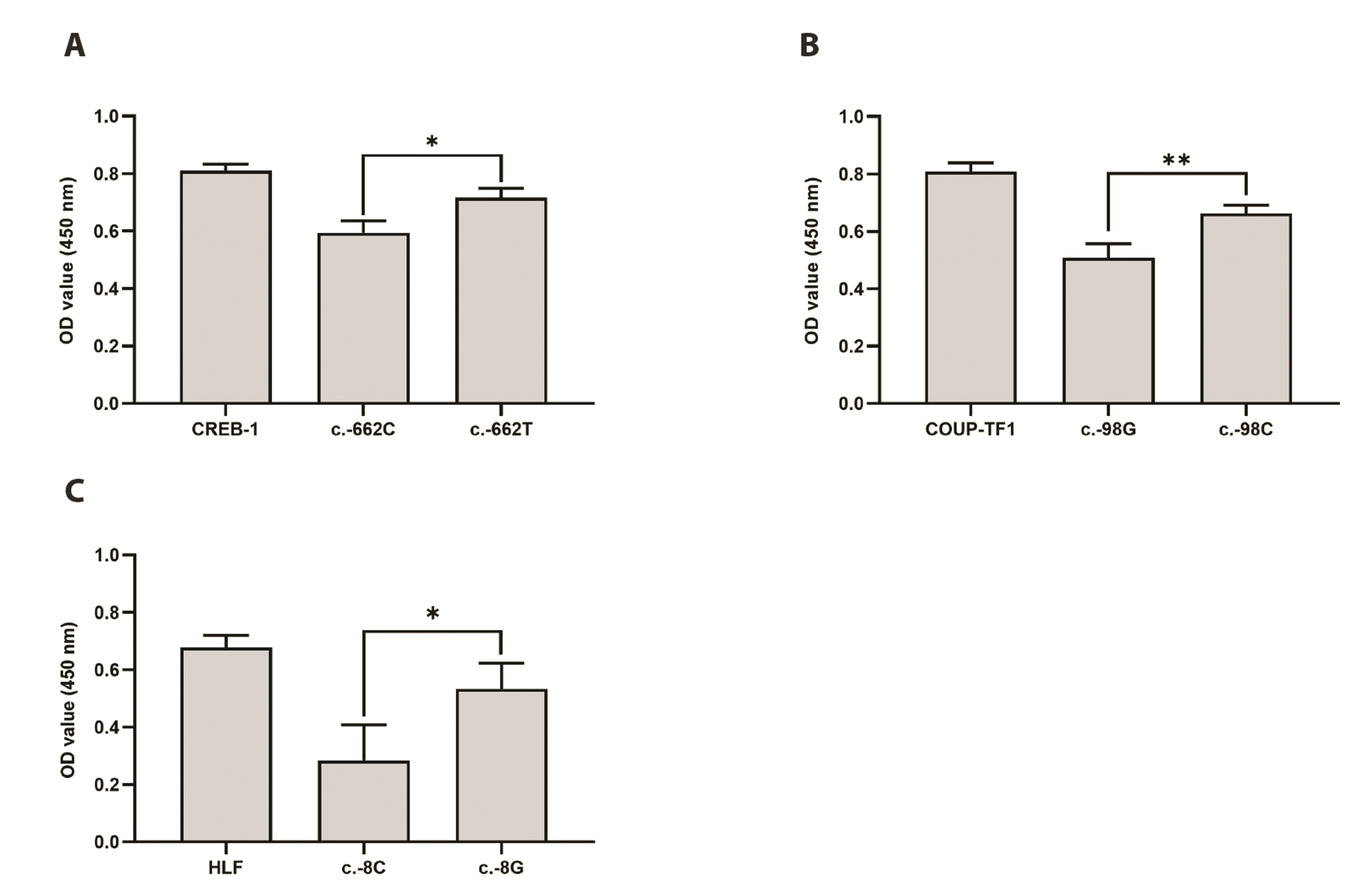
- Solute carrier 40A1 (SLC40A1) encodes ferroportin, which is the only known transmembrane protein that exports elemental iron from mammalian cells and is essential for iron homeostasis. Mutations in SLC40A1 are associated with ironoverload disorders. In addition to ferroportin diseases, SLC40A1 expression is downregulated in various cancer types. Despite the clinical significance of the SLC40A1 transporter, only a few studies have investigated genetic variants in SLC40A1. The present study was performed to identify genetic variations in the SLC40A1 promoter and functionally characterize each variant using in vitro assays. We investigated four haplotypes and five variants in the SLC40A1 promoter. We observed that haplotype 3 (H3) had significantly lower promoter activity than H1, whereas the activity of H4 was significantly higher than that of H1. Luciferase activity of H2 was comparable to that of H1. In addition, four variants of SLC40A1, c.-1355G>C, c.-662C>T, c.-98G>C, and c.-8C>G, showed significantly increased luciferase activity compared to the wild type (WT), whereas c.-750G>A showed significantly decreased luciferase activity compared to the WT. Three transcription factors, cAMP response element-binding protein-1 (CREB-1), chicken ovalbumin upstream promoter transcription factor 1, and hepatic leukemia factor (HLF), were predicted to bind to the promoter regions of SLC40A1 near c.-662C>T, c.-98G>C, and c.-8C>G, respectively. Among these, CREB-1 and HLF bound more strongly to the variant sequences than to the WT and functioned as activators of SLC40A1 transcription. Collectively, our findings indicate that the two SLC40A1 promoter haplotypes affect SLC40A1 transcription, which is regulated by CREB-1 and HLF.
Keyword
Figure
Reference
-
1. Ravasi G, Pelucchi S, Russo A, Mariani R, Piperno A. 2020; Ferroportin disease: a novel SLC40A1 mutation. Dig Liver Dis. 52:688–690. DOI: 10.1016/j.dld.2020.03.013. PMID: 32360131.
Article2. Moreno-Carralero MI, Muñoz-Muñoz JA, Cuadrado-Grande N, López-Rodríguez R, José Hernández-Alfaro M, Enríquez-de-Salamanca R, Méndez M, Morán-Jiménez MJ. del-Castillo-Rueda A. 2014; A novel mutation in the SLC40A1 gene associated with reduced iron export in vitro. Am J Hematol. 89:689–694. DOI: 10.1002/ajh.23714. PMID: 24644245.3. Détivaud L, Island ML, Jouanolle AM, Ropert M, Bardou-Jacquet E, Le Lan C, Mosser A, Leroyer P, Deugnier Y, David V, Brissot P, Loréal O. 2013; Ferroportin diseases: functional studies, a link between genetic and clinical phenotype. Hum Mutat. 34:1529–1536. DOI: 10.1002/humu.22396. PMID: 23943237.
Article4. Mayr R, Griffiths WJ, Hermann M, McFarlane I, Halsall DJ, Finkenstedt A, Douds A, Davies SE, Janecke AR, Vogel W, Cox TM, Zoller H. 2011; Identification of mutations in SLC40A1 that affect ferroportin function and phenotype of human ferroportin iron overload. Gastroenterology. 140:2056–2063. 2063.e1. DOI: 10.1053/j.gastro.2011.02.064. PMID: 21396368.
Article5. Lelièvre P, Sancey L, Coll JL, Deniaud A, Busser B. 2020; Iron dysregulation in human cancer: altered metabolism, biomarkers for diagnosis, prognosis, monitoring and rationale for therapy. Cancers (Basel). 12:3524. DOI: 10.3390/cancers12123524. PMID: 33255972. PMCID: PMC7761132.
Article6. Xue D, Zhou CX, Shi YB, Lu H, He XZ. 2015; Decreased expression of ferroportin in prostate cancer. Oncol Lett. 10:913–916. Erratum in: Oncol Lett. 2021;21:257. DOI: 10.3892/ol.2015.3363. PMID: 26622594. PMCID: PMC4509077.
Article7. Deng Z, Manz DH, Torti SV, Torti FM. 2019; Effects of ferroportin-mediated iron depletion in cells representative of different histological subtypes of prostate cancer. Antioxid Redox Signal. 30:1043–1061. DOI: 10.1089/ars.2017.7023. PMID: 29061069. PMCID: PMC6354616.
Article8. Pinnix ZK, Miller LD, Wang W, D'Agostino R Jr, Kute T, Willingham MC, Hatcher H, Tesfay L, Sui G, Di X, Torti SV, Torti FM. 2010; Ferroportin and iron regulation in breast cancer progression and prognosis. Sci Transl Med. 2:43ra56. Erratum in: Sci Transl Med. 2010;2:46er1. DOI: 10.1126/scitranslmed.3001127. PMID: 20686179. PMCID: PMC3734848.
Article9. Deng L, Huang L, Guo Q, Shi X, Xu K. 2017; CREB1 and Smad3 mediate TGF-β3-induced Smad7 expression in rat hepatic stellate cells. Mol Med Rep. 16:8455–8462. DOI: 10.3892/mmr.2017.7654. PMID: 28983617.
Article10. Chen HC, Byrd JC, Muthusamy N. 2006; Differential role for cyclic AMP response element binding protein-1 in multiple stages of B cell development, differentiation, and survival. J Immunol. 176:2208–2218. DOI: 10.4049/jimmunol.176.4.2208. PMID: 16455977.
Article11. Barral S, Reitz C, Small SA, Mayeux R. 2014; Genetic variants in a 'cAMP element binding protein' (CREB)-dependent histone acetylation pathway influence memory performance in cognitively healthy elderly individuals. Neurobiol Aging. 35:2881.e7–2881.e10. DOI: 10.1016/j.neurobiolaging.2014.06.024. PMID: 25150575. PMCID: PMC4253058.
Article12. Wang P, Deng L, Zhuang C, Cheng C, Xu K. 2016; p-CREB-1 promotes hepatic fibrosis through the transactivation of transforming growth factor-β1 expression in rats. Int J Mol Med. 38:521–528. DOI: 10.3892/ijmm.2016.2630. PMID: 27279449.
Article13. Xin ZC, Hu HW, Lao ZH, Zhu LQ, Biskup E, Zhang HW. 2020; p-CREB-1 at Ser 133 is a potential marker for breast cancer. Eur Rev Med Pharmacol Sci. 24:11628–11638.14. Xiao X, Zhang C, Grigoroiu-Serbanescu M, Wang L, Li L, Zhou D, Yuan TF, Wang C, Chang H, Wu Y, Li Y, Wu DD, Yao YG, Li M. 2018; The cAMP responsive element-binding (CREB)-1 gene increases risk of major psychiatric disorders. Mol Psychiatry. 23:1957–1967. DOI: 10.1038/mp.2017.243. PMID: 29158582.
Article15. Nabokina SM, Ramos MB, Valle JE, Said HM. 2015; Regulation of basal promoter activity of the human thiamine pyrophosphate transporter SLC44A4 in human intestinal epithelial cells. Am J Physiol Cell Physiol. 308:C750–C757. Erratum in: Am J Physiol Cell Physiol. 2017;313:C473. DOI: 10.1152/ajpcell.00381.2014. PMID: 25715703. PMCID: PMC4420793.16. Teng X, Liu YY, Teng W, Brent GA. 2018; COUP-TF1 modulates thyroid hormone action in an embryonic stem-cell model of cortical pyramidal neuronal differentiation. Thyroid. 28:667–678. DOI: 10.1089/thy.2017.0256. PMID: 29205104. PMCID: PMC5952340.
Article17. Fischer SF, Schmidt K, Fiedler N, Glebe D, Schüttler C, Sun J, Gerlich WH, Repp R, Schaefer S. 2006; Genotype-dependent activation or repression of HBV enhancer II by transcription factor COUP-TF1. World J Gastroenterol. 12:6054–6058. DOI: 10.3748/wjg.v12.i37.6054. PMID: 17009409. PMCID: PMC4124418.
Article18. Yamaguchi H, Zhou C, Lin SC, Durand B, Tsai SY, Tsai MJ. 2004; The nuclear orphan receptor COUP-TFI is important for differentiation of oligodendrocytes. Dev Biol. 266:238–251. DOI: 10.1016/j.ydbio.2003.10.038. PMID: 14738874.
Article19. Rodriguez-Tirado C, Kale N, Carlini MJ, Shrivastava N, Rodrigues AA, Khalil BD, Bravo-Cordero JJ, Hong Y, Alexander M, Ji J, Behbod F, Sosa MS. 2022; NR2F1 is a barrier to dissemination of early-stage breast cancer cells. Cancer Res. 82:2313–2326. DOI: 10.1158/0008-5472.CAN-21-4145. PMID: 35471456. PMCID: PMC9203932.
Article20. Perets R, Kaplan T, Stein I, Hidas G, Tayeb S, Avraham E, Ben-Neriah Y, Simon I, Pikarsky E. 2012; Genome-wide analysis of androgen receptor targets reveals COUP-TF1 as a novel player in human prostate cancer. PLoS One. 7:e46467. DOI: 10.1371/journal.pone.0046467. PMID: 23056316. PMCID: PMC3464259.
Article21. Hunger SP, Ohyashiki K, Toyama K, Cleary ML. 1992; Hlf, a novel hepatic bZIP protein, shows altered DNA-binding properties following fusion to E2A in t(17;19) acute lymphoblastic leukemia. Genes Dev. 6:1608–1620. DOI: 10.1101/gad.6.9.1608. PMID: 1516826.
Article22. Wahlestedt M, Ladopoulos V, Hidalgo I, Sanchez Castillo M, Hannah R, Säwén P, Wan H, Dudenhöffer-Pfeifer M, Magnusson M, Norddahl GL, Göttgens B, Bryder D. 2017; Critical modulation of hematopoietic lineage fate by hepatic leukemia factor. Cell Rep. 21:2251–2263. DOI: 10.1016/j.celrep.2017.10.112. PMID: 29166614. PMCID: PMC5714592.
Article23. Waters KM, Sontag RL, Weber TJ. 2013; Hepatic leukemia factor promotes resistance to cell death: implications for therapeutics and chronotherapy. Toxicol Appl Pharmacol. 268:141–148. DOI: 10.1016/j.taap.2013.01.031. PMID: 23415677.
Article24. Xiang DM, Sun W, Zhou T, Zhang C, Cheng Z, Li SC, Jiang W, Wang R, Fu G, Cui X, Hou G, Jin GZ, Li H, Hou C, Liu H, Wang H, Ding J. 2019; Oncofetal HLF transactivates c-Jun to promote hepatocellular carcinoma development and sorafenib resistance. Gut. 68:1858–1871. DOI: 10.1136/gutjnl-2018-317440. PMID: 31118247.
Article25. Xiang DM, Sun W, Ning BF, Zhou TF, Li XF, Zhong W, Cheng Z, Xia MY, Wang X, Deng X, Wang W, Li HY, Cui XL, Li SC, Wu B, Xie WF, Wang HY, Ding J. 2018; The HLF/IL-6/STAT3 feedforward circuit drives hepatic stellate cell activation to promote liver fibrosis. Gut. 67:1704–1715. DOI: 10.1136/gutjnl-2016-313392. PMID: 28754776.
Article26. Inukai T, Hirose K, Inaba T, Kurosawa H, Hama A, Inada H, Chin M, Nagatoshi Y, Ohtsuka Y, Oda M, Goto H, Endo M, Morimoto A, Imaizumi M, Kawamura N, Miyajima Y, Ohtake M, Miyaji R, Saito M, Tawa A, et al. 2007; Hypercalcemia in childhood acute lymphoblastic leukemia: frequent implication of parathyroid hormone-related peptide and E2A-HLF from translocation 17;19. Leukemia. 21:288–296. DOI: 10.1038/sj.leu.2404496. PMID: 17183364.
Article27. Liu Q, Ge H, Liu P, Li Y. 2021; High Hepatic leukemia factor expression indicates a favorable survival in glioma patients. Medicine (Baltimore). 100:e23980. DOI: 10.1097/MD.0000000000023980. PMID: 33578515. PMCID: PMC7886392.
Article28. Kim BM, Song HS, Kim JY, Kwon EY, Ha SY, Kim M, Choi JH. 2022; Functional characterization of ABCA4 genetic variants related to Stargardt disease. Sci Rep. 12:22282. DOI: 10.1038/s41598-022-26912-6. PMID: 36566289. PMCID: PMC9790013.
Article29. Vivot A, Boutron I, Ravaud P, Porcher R. 2015; Guidance for pharmacogenomic biomarker testing in labels of FDA-approved drugs. Genet Med. 17:733–738. DOI: 10.1038/gim.2014.181. PMID: 25521333.
Article30. Cooper-DeHoff RM, Niemi M, Ramsey LB, Luzum JA, Tarkiainen EK, Straka RJ, Gong L, Tuteja S, Wilke RA, Wadelius M, Larson EA, Roden DM, Klein TE, Yee SW, Krauss RM, Turner RM, Palaniappan L, Gaedigk A, Giacomini KM, Caudle KE, et al. 2022; The clinical pharmacogenetics implementation consortium guideline for SLCO1B1, ABCG2, and CYP2C9 genotypes and statin-associated musculoskeletal symptoms. Clin Pharmacol Ther. 111:1007–1021. DOI: 10.1002/cpt.2557. PMID: 35152405. PMCID: PMC9035072.31. Peltekova VD, Wintle RF, Rubin LA, Amos CI, Huang Q, Gu X, Newman B, Van Oene M, Cescon D, Greenberg G, Griffiths AM, St George-Hyslop PH, Siminovitch KA. 2004; Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat Genet. 36:471–475. DOI: 10.1038/ng1339. PMID: 15107849.
Article32. Leung E, Hong J, Fraser AG, Merriman TR, Vishnu P, Krissansen GW. 2006; Polymorphisms in the organic cation transporter genes SLC22A4 and SLC22A5 and Crohn's disease in a New Zealand Caucasian cohort. Immunol Cell Biol. 84:233–236. DOI: 10.1111/j.1440-1711.2006.01423.x. PMID: 16519742.
Article33. Martínez A, Martín MC, Mendoza JL, Taxonera C, Díaz-Rubio M, de la Concha EG, Urcelay E. 2006; Association of the organic cation transporter OCTN genes with Crohn's disease in the Spanish population. Eur J Hum Genet. 14:222–226. DOI: 10.1038/sj.ejhg.5201529. PMID: 16333318.
Article34. Tomer G, Wetzler G, Keddache M, Denson LA. 2009; Polymorphisms in the IBD5 locus are associated with Crohn disease in pediatric Ashkenazi Jewish patients. J Pediatr Gastroenterol Nutr. 48:531–537. DOI: 10.1097/MPG.0b013e318183138a. PMID: 19412005.
Article35. Jung ES, Park HJ, Kong KA, Choi JH, Cheon JH. 2017; Association study between OCTN1 functional haplotypes and Crohn's disease in a Korean population. Korean J Physiol Pharmacol. 21:11–17. DOI: 10.4196/kjpp.2017.21.1.11. PMID: 28066136. PMCID: PMC5214902.
Article36. Park HJ, Jung ES, Kong KA, Park EM, Cheon JH, Choi JH. 2016; Identification of OCTN2 variants and their association with phenotypes of Crohn's disease in a Korean population. Sci Rep. 6:22887. DOI: 10.1038/srep22887. PMID: 26965072. PMCID: PMC4786794.
Article37. Brookes MJ, Hughes S, Turner FE, Reynolds G, Sharma N, Ismail T, Berx G, McKie AT, Hotchin N, Anderson GJ, Iqbal T, Tselepis C. 2006; Modulation of iron transport proteins in human colorectal carcinogenesis. Gut. 55:1449–1460. DOI: 10.1136/gut.2006.094060. PMID: 16641131. PMCID: PMC1856421.
Article38. Liang B, Zhou C, Cui S, Lu H, Xu R, Xue D, Zou S, He X. 2021; Upregulation of miR-18a-5p promotes the proliferation of prostate cancer via inhibiting the expression of SLC40A1. Pathol Res Pract. 224:153448. DOI: 10.1016/j.prp.2021.153448. PMID: 34098197.
Article39. Wu J, Zhang L, Wu S, Yi X, Liu Z. 2020; miR-194-5p inhibits SLC40A1 expression to induce cisplatin resistance in ovarian cancer. Pathol Res Pract. 216:152979. DOI: 10.1016/j.prp.2020.152979. PMID: 32534701.
Article40. Gasparetto M, Pei S, Minhajuddin M, Stevens B, Smith CA, Seligman P. 2019; Low ferroportin expression in AML is correlated with good risk cytogenetics, improved outcomes and increased sensitivity to chemotherapy. Leuk Res. 80:1–10. DOI: 10.1016/j.leukres.2019.02.011. PMID: 30852438.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Identification and Functional Characterization of Novel Genetic Variations in the OCTN1 Promoter
- Identification of Novel Genetic Variations in the Proximal Promoter of the Human Transporter, OCT2
- The DPE, a core promoter element for transcription by RNA polymerase II
- Identification and Functional Characterization of ST3GAL5 and ST8SIA1 Variants in Patients with Thyroid-Associated Ophthalmopathy
- The upstream sequence of Mycobacterium leprae 18-kDa gene confers transcription repression activity in orientation-independent manner