Brain Tumor Res Treat.
2024 Jan;12(1):23-39. 10.14791/btrt.2023.0036.
Comprehensive Molecular Genetic Analysis in Glioma Patients by Next Generation Sequencing
- Affiliations
-
- 1Departments of Hospital Pathology , Seoul St. Mary’s Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 2Departments of Neurosurgery, Seoul St. Mary’s Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea
- KMID: 2552336
- DOI: http://doi.org/10.14791/btrt.2023.0036
Abstract
- Background
Glioma is caused by multiple genomic alterations. The evolving classification of glio- mas emphasizes the significance of molecular testing. Next generation sequencing (NGS) offers the assessment of parallel combinations of multiple genetic alterations and identifying actionable mutations that guide treatment. This study comprehensively analyzed glioma patients using multi-gene NGS panels, providing powerful insights to inform diagnostic classification and targeted therapies.
Methods
We conducted a targeted panel-based NGS analysis on formalin-fixed and paraffin- embedded nucleic acids extracted from a total of 147 glioma patients. These samples underwent amplicon capture-based library preparation and sequenced using the Oncomine Comprehensive Assay platform. The resulting sequencing data were then analyzed using the bioinformatics tools.
Results
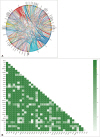
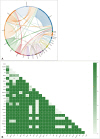
A total of 301 mutations, were found in 132 out of 147 tumors (89.8%). These muta- tions were in 68 different genes. In 62 tumor samples (42.2%), copy number variations (CNVs) with gene amplifications occurred in 25 genes. Moreover, 25 tumor samples (17.0%) showed gene fusions in 6 genes and intragenic deletion in a gene. Our analysis identified actionable targets in several genes, including 11 with mutations, 8 with CNVs, and 3 with gene fusions and intragenic deletion. These findings could impact FDA-approved therapies, NCCN guideline-based treatments, and clinical trials.
Conclusion
We analyzed precisely diagnosing the classification of gliomas, detailing the frequency and co-occurrence of genetic alterations and identifying genetic alterations with potential therapeutic targets by NGS-based molecular analysis. The high-throughput NGS analysis is an efficient and powerful tool to comprehensively support molecular testing in neurooncology.
Figure
Reference
-
1. Molinaro AM, Taylor JW, Wiencke JK, Wrensch MR. Genetic and molecular epidemiology of adult diffuse glioma. Nat Rev Neurol. 2019; 15:405–417. PMID: 31227792.
Article2. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016; 131:803–820. PMID: 27157931.
Article3. Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol. 2021; 23:1231–1251. PMID: 34185076.
Article4. Lorenz J, Rothhammer-Hampl T, Zoubaa S, Bumes E, PukropT , Kolbl O, et al. A comprehensive DNA panel next generation sequencing approach supporting diagnostics and therapy prediction inneurooncology. Acta Neuropathol Commun. 2020; 8:124. PMID: 32758285.
Article5. Tirrò E, Massimino M, Broggi G, Romano C, Minasi S, Gianno F, et al. Custom DNA-based NGS panel for the molecular characterization of patients with diffuse gliomas: diagnostic and therapeutic applications. Front Oncol. 2022; 12:861078. PMID: 35372034.
Article6. Śledzińska P, Bebyn M, Szczerba E, Furtak J, Harat M, Olszewska N, et al. Glioma 2021 WHO classification: the superiority of NGS Over IHC in routine diagnostics. Mol Diagn Ther. 2022; 26:699–713. PMID: 36053463.7. Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005; 109:93–108. PMID: 15685439.8. Sonoda Y, Kumabe T, Nakamura T, Saito R, Kanamori M, Yamashita Y, et al. Analysis of IDH1 and IDH2 mutations in Japanese glioma patients. Cancer Sci. 2009; 100:1996–1998. PMID: 19765000.9. Yuan Y, Qi C, Maling G, Xiang W, Yanhui L, Ruofei L, et al. TERT mutation in glioma: frequency, prognosis and risk. J Clin Neurosci. 2016; 26:57–62. PMID: 26765760.
Article10. Powter B, Jeffreys SA, Sareen H, Cooper A, Brungs D, Po J, et al. Human TERT promoter mutations as a prognostic biomarker in glioma. J Cancer Res Clin Oncol. 2021; 147:1007–1017. PMID: 33547950.
Article11. Hoogstrate Y, Vallentgoed W, Kros JM, de Heer I, de Wit M, Eoli M, et al. EGFR mutations are associated with response to depatux-m in combination with temozolomide and result in a receptor that is hypersensitive to ligand. Neurooncol Adv. 2019; 2:vdz051. PMID: 32642719.
Article12. Pappula AL, Rasheed S, Mirzaei G, Petreaca RC, Bouley RA. A genome-wide profiling of glioma patients with an IDH1 mutation using the Catalogue of Somatic Mutations in Cancer Database. Cancers (Basel). 2021; 13:4299. PMID: 34503108.
Article13. Cahill DP, Louis DN, Cairncross JG. Molecular background of oligodendroglioma: 1p/19q, IDH, TERT, CIC and FUBP1. CNS Oncol. 2015; 4:287–294. PMID: 26545048.
Article14. Parsons DW, Jones S, Zhang X, Lin JCH, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008; 321:1807–1812. PMID: 18772396.
Article15. Fontebasso AM, Papillon-Cavanagh S, Schwartzentruber J, Nikbakht H, Gerges N, Fiset PO, et al. Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nat Genet. 2014; 46:462–466. PMID: 24705250.
Article16. Erira A, Velandia F, Penagos J, Zubieta C, Arboleda G. Differential regulation of the EGFR/PI3K/AKT/PTEN pathway between low- and high-grade gliomas. Brain Sci. 2021; 11:1655. PMID: 34942957.17. Zeng C, Wang J, Li M, Wang H, Lou F, Cao S, et al. Comprehensive molecular characterization of Chinese patients with glioma by extensive next-generation sequencing panel analysis. Cancer Manag Res. 2021; 13:3573–3588. PMID: 33953611.
Article18. González-Tablas M, Arandia D, Jara-Acevedo M, Otero Á, Vital AL, Prieto C, et al. Heterogeneous EGFR, CDK4, MDM4, and PDGFRA gene expression profiles in primary GBM: no association with patient survival. Cancers (Basel). 2020; 12:231. PMID: 31963499.
Article19. Furgason JM, Koncar RF, Michelhaugh SK, Sarkar FH, Mittal S, Sloan AE, et al. Whole genome sequence analysis links chromothripsis to EGFR, MDM2, MDM4, and CDK4 amplification in glioblastoma. Oncoscience. 2015; 2:618–628. PMID: 26328271.
Article20. An Z, Aksoy O, Zheng T, Fan QW, Weiss WA. Epidermal growth factor receptor and EGFRvIII in glioblastoma: signaling pathways and targeted therapies. Oncogene. 2018; 37:1561–1575. PMID: 29321659.
Article21. Lassman AB, Aldape KD, Ansell PJ, Bain E, Curran WJ, Eoli M, et al. Epidermal growth factor receptor (EGFR) amplification rates observed in screening patients for randomized trials in glioblastoma. J Neurooncol. 2019; 144:205–210. PMID: 31273577.
Article22. Zwick E, Bange J, Ullrich A. Receptor tyrosine kinase signalling as a target for cancer intervention strategies. Endocr Relat Cancer. 2001; 8:161–173. PMID: 11566607.
Article23. Garima G, Thanvi S, Singh A, Verma V. Epidermal growth factor receptor variant III mutation, an emerging molecular marker in glioblastoma multiforme patients: a single institution study on the Indian population. Cureus. 2022; 14:e26412. PMID: 35911278.
Article24. Shah N, Lankerovich M, Lee H, Yoon JG, Schroeder B, Foltz G. Exploration of the gene fusion landscape of glioblastoma using transcriptome sequencing and copy number data. BMC Genomics. 2013; 14:818. PMID: 24261984.
Article25. Frattini V, Pagnotta SM, Tala Fan JJ, Russo MV, Lee SB, Garofano L, et al. A metabolic function of FGFR3-TACC3 gene fusions in cancer. Nature. 2018; 553:222–227. PMID: 29323298.
Article26. Ferguson SD, Zhou S, Huse JT, de Groot JF, Xiu J, Subramaniam DS, et al. Targetable gene fusions associate with the IDH wild-type astrocytic lineage in adult gliomas. J Neuropathol Exp Neurol. 2018; 77:437–442. PMID: 29718398.
Article27. Bao ZS, Chen HM, Yang MY, Zhang CB, Yu K, Ye WL, et al. RNA-seq of 272 gliomas revealed a novel, recurrent PTPRZ1-MET fusion transcript in secondary glioblastomas. Genome Res. 2014; 24:1765–1773. PMID: 25135958.
Article28. Mrowczynski OD, Payne R, Pu C, Greiner R, Rizk E. A unique case of a high-grade neuroepithelial tumor with EML4-ALK fusion in a five-month-old. Cureus. 2020; 12:e8654. PMID: 32685319.
Article29. Pajtler KW, Witt H, Sill M, Jones DT, Hovestadt V, Kratochwil F, et al. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell. 2015; 27:728–743. PMID: 25965575.
Article30. Padovan M, Maccari M, Bosio A, De Toni C, Vizzaccaro S, Cestonaro I, et al. Actionable molecular alterations in newly diagnosed and recurrent IDH1/2 wild-type glioblastoma patients and therapeutic implications: a large mono-institutional experience using extensive next-generation sequencing analysis. Eur J Cancer. 2023; 191:112959. PMID: 37481865.
Article31. Ministry of Food and Drug Safety. Search for clinical trial information [Internet]. Cheongju: Ministry of Food and Drug Safety;2023. Accessed August 1, 2023. at: https://nedrug.mfds.go.kr/searchClinic.32. Mellinghoff IK, van den Bent MJ, Blumenthal DT, Touat M, Peters KB, Clarke J, et al. Vorasidenib in IDH1- or IDH2-mutant low-grade glioma. N Engl J Med. 2023; 389:589–601. PMID: 37272516.
Article33. Mellinghoff IK, van den Bent MJ, Blumenthal DT, Touat M, Peters KB, Clarke JL, et al. INDIGO: A global, randomized, double-blinded, phase 3 study of vorasidenib versus placebo in patients with residual or recurrent grade 2 glioma with an IDH1/2 mutation. J Clin Oncol. 2023; 41(suppl 17):LBA1.34. Arbour G, Ellezam B, Weil AG, Cayrol R, Vanan MI, Coltin H, et al. Upfront BRAF/MEK inhibitors for treatment of high-grade glioma: a case report and review of the literature. Neurooncol Adv. 2022; 4:vdac174. PMID: 36567957.
Article35. Olympios N, Gilard V, Marguet F, Clatot F, Di Fiore F, Fontanilles M. TERT promoter alterations in glioblastoma: a systematic review. Cancers (Basel). 2021; 13:1147. PMID: 33800183.
Article36. Di Stefano AL, Picca A, Saragoussi E, Bielle F, Ducray F, Villa C, et al. Clinical, molecular, and radiomic profile of gliomas with FGFR3-TACC3 fusions. Neuro Oncol. 2020; 22:1614–1624. PMID: 32413119.
Article37. Wen PY, de Groot JF, Battiste J, Goldlust SA, Garner JS, Friend J, et al. Paxalisib in patients with newly diagnosed glioblastoma with unmethylated MGMT promoter status: final phase 2 study results. J Clin Oncol. 2022; 40(16_suppl):2047.
Article38. Tiu C, Biondo A, Welsh LC, Jones TL, Zachariou A, Prout T, et al. Results of the glioblastoma multiforme (GBM) cohort of phase 1 trial Ice-CAP (NCT03673787): preliminary evidence of antitumour activity of Ipatasertib (Ipa) and Atezolizumab (A) in patients (pts) with PTEN loss. Cancer Res. 2021; 81(13_Supplement):CT120.39. Alexandru O, Horescu C, Sevastre AS, Cioc CE, Baloi C, Oprita A, et al. Receptor tyrosine kinase targeting in glioblastoma: performance, limitations and future approaches. Contemp Oncol (Pozn). 2020; 24:55–66. PMID: 32514239.
Article40. Bolcaen J, Nair S, Driver CHS, Boshomane TMG, Ebenhan T, Vandevoorde C. Novel receptor tyrosine kinase pathway inhibitors for targeted radionuclide therapy of glioblastoma. Pharmaceuticals (Basel). 2021; 14:626. PMID: 34209513.
Article41. Lombardi G, Caccese M, Padovan M, Cerretti G, Pintacuda G, Manara R, et al. Regorafenib in recurrent glioblastoma patients: a large and monocentric real-life study. Cancers (Basel). 2021; 13:4731. PMID: 34572958.
Article42. Sepúlveda-Sánchez JM, Vaz MÁ, Balañá C, Gil-Gil M, Reynés G, Gallego Ó, et al. Phase II trial of dacomitinib, a pan-human EGFR tyrosine kinase inhibitor, in recurrent glioblastoma patients with EGFR amplification. Neuro Oncol. 2017; 19:1522–1531. PMID: 28575464.
Article43. Sepúlveda JM, Sánchez-Gómez P, Vaz Salgado MÁ, Gargini R, Balañá C. Dacomitinib: an investigational drug for the treatment of glioblastoma. Expert Opin Investig Drugs. 2018; 27:823–829.
Article44. Byeon S, Hong JY, Lee J, Nam DH, Park SH, Park JO, et al. Use of gefitinib in EGFR-amplified refractory solid tumors: an open-label, single-arm. single-center prospective pilot study. Target Oncol. 2020; 15:185–192. PMID: 32107712.
Article45. O’Rourke DM, Nasrallah MP, Desai A, Melenhorst JJ, Mansfield K, Morrissette JJ, et al. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci Trans Med. 2017; 9:eaaa0984.
Article46. Durgin JS, Henderson F Jr, Nasrallah MP, Mohan S, Wang S, Lacey SF, et al. Case report: prolonged survival following EGFRvIII CAR T cell treatment for recurrent glioblastoma. Front Oncol. 2021; 11:669071. PMID: 34026647.
Article47. Szklener K, Mazurek M, Wieteska M, Wacławska M, Bilski M, Mańdziuk S. New directions in the therapy of glioblastoma. Cancers (Basel). 2022; 31:14.
Article48. Bao Z, Li S, Wang L, Zhang B, Zhang P, Shi H, et al. PTPRZ1-METFUsion GENe (ZM-FUGEN) trial: study protocol for a multicentric, randomized, open-label phase II/III trial. Chin Neurosurg J. 2023; 9:21. PMID: 37443050.
Article49. Salloum R, Huang J, Stewart CF, Fuller C, Smolarek T, Lenzen A, et al. TRLS-11. A phase 1 study of savolitinib in recurrent, progressive, or refractory medulloblastoma, high grade glioma, diffuse intrinsic pontine glioma, and central nervous system (CNS) tumors harboring MET aberrations: a pediatric brain tumor consortium trial. Neuro Oncol. 2023; 25(Suppl 1):i81.50. Hong D, Catenacci D, Bazhenova L, Cho BC, Ponz-Sarvise M, Heist R, et al. Preliminary interim data of elzovantinib (TPX-0022), a novel inhibitor of MET/SRC/CSF1R, in patients with advanced solid tumors harboring genetic alterations in MET: update from the phase 1 SHIELD-1 trial. Mol Cancer Ther. 2021; 20(12_Supplement):P225.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Application of Next Generation Sequencing Upon the Molecular Genetic Diagnosis of Deafness
- Principles of Genetic Counseling in the Era of Next-Generation Sequencing
- Molecular genetic decoding of malformations of cortical development
- Recent Advances in the Clinical Application of Next-Generation Sequencing
- The molecular pathophysiology of vascular anomalies: Genomic research