J Korean Med Sci.
2024 Feb;39(5):e47. 10.3346/jkms.2024.39.e47.
Validation of the Framingham Diabetes Risk Model Using Community-Based KoGES Data
- Affiliations
-
- 1Clinical Trial Center, Ewha Womans University Mokdong Hospital, Seoul, Korea
- 2Department of Preventive Medicine, College of Medicine, Ewha Womans University, Seoul, Korea
- 3Graduate Program in System Health Science and Engineering, Ewha Womans University, Seoul, Korea
- 4Department of Internal Medicine, College of Medicine, Ewha Womans University, Seoul, Korea
- KMID: 2551977
- DOI: http://doi.org/10.3346/jkms.2024.39.e47
Abstract
- Background
An 8-year prediction of the Framingham Diabetes Risk Model (FDRM) was proposed, but the predictor has a gap with current clinical standards. Therefore, we evaluated the validity of the original FDRM in Korean population data, developed a modified FDRM by redefining the predictors based on current knowledge, and evaluated the internal and external validity.
Methods
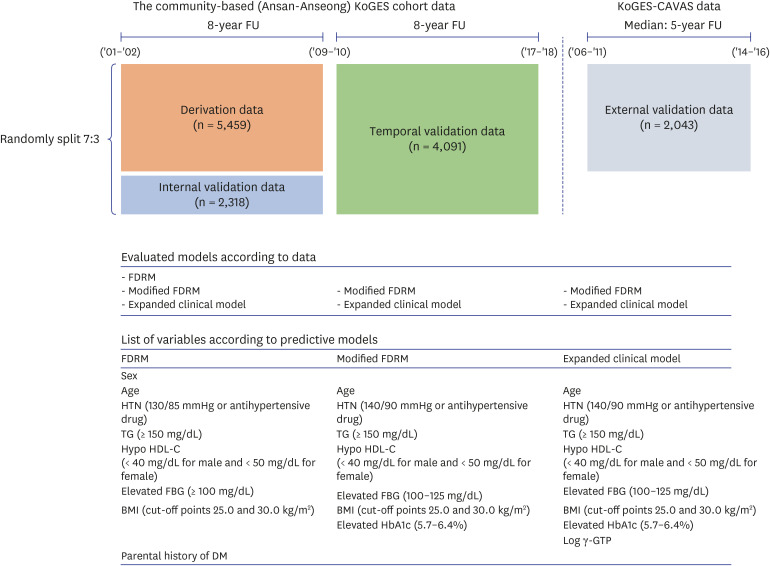
Using data from a community-based cohort in Korea (n = 5,409), we calculated the probability of diabetes through FDRM, and developed a modified FDRM based on modified definitions of hypertension (HTN) and diabetes. We also added clinical features related to diabetes to the predictive model. Model performance was evaluated and compared by area under the curve (AUC).
Results
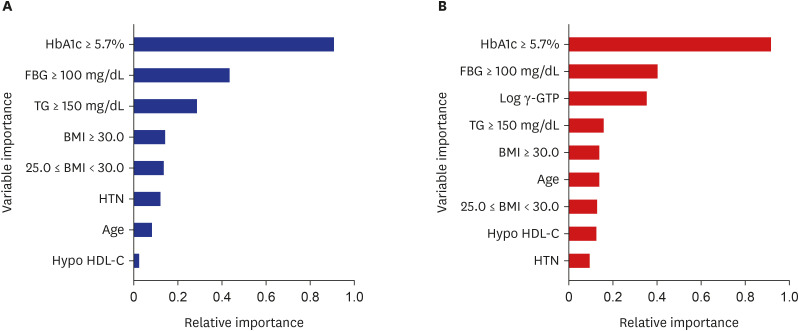
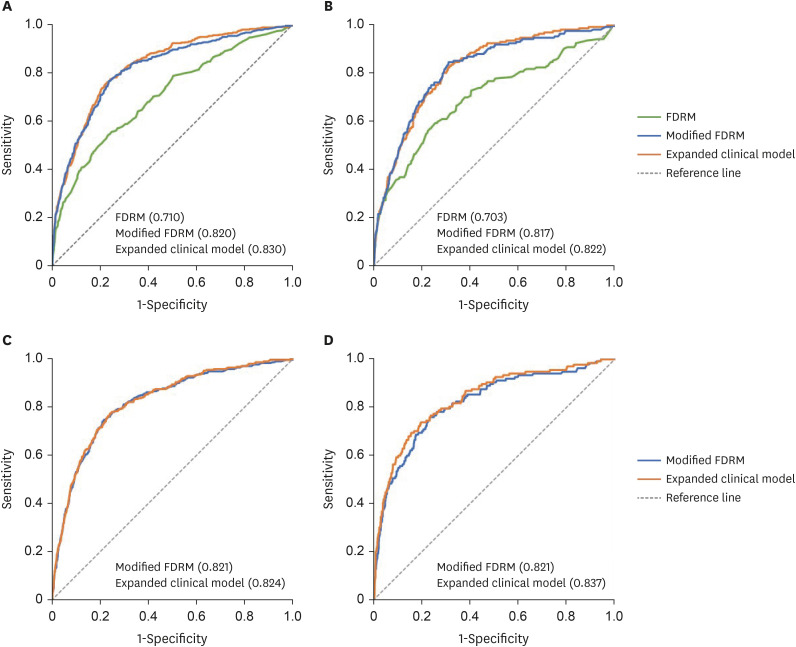
During the 8-year follow-up, the cumulative incidence of diabetes was 8.5%. The modified FDRM consisted of age, obesity, HTN, hypo-high-density lipoprotein cholesterol, elevated triglyceride, fasting glucose, and hemoglobin A1c. The expanded clinical model added γ-glutamyl transpeptidase to the modified FDRM. The FDRM showed an estimated AUC of 0.71, and the model's performance improved to an AUC of 0.82 after applying the redefined predictor. Adding clinical features (AUC = 0.83) to the modified FDRM further improved in discrimination, but this was not maintained in the validation data set. External validation was evaluated on population-based cohort data and both modified models performed well, with AUC above 0.82.
Conclusion
The performance of FDRM in the Korean population was found to be acceptable for predicting diabetes, but it was improved when corrected with redefined predictors. The validity of the modified model needs to be further evaluated.
Figure
Reference
-
1. World Health Organization, Chronic Respiratory Diseases and Arthritis Team. Screening for type 2 diabetes: report of a World Health Organization and International Diabetes Federation meeting. Updated 2003. Accessed March 1, 2023. https://apps.who.int/iris/handle/10665/68614 .2. International Diabetes Federation. IDF Diabetes Atlas. 10th ed. Brussels, Belgium: International Diabetes Federation;2021.3. Herman WH, Ye W, Griffin SJ, Simmons RK, Davies MJ, Khunti K, et al. Early detection and treatment of type 2 diabetes reduce cardiovascular morbidity and mortality: a simulation of the results of the Anglo-Danish-Dutch Study of Intensive Treatment in People with Screen-Detected Diabetes in Primary Care (ADDITION-Europe). Diabetes Care. 2015; 38(8):1449–1455. PMID: 25986661.4. Fregoso-Aparicio L, Noguez J, Montesinos L, García-García JA. Machine learning and deep learning predictive models for type 2 diabetes: a systematic review. Diabetol Metab Syndr. 2021; 13(1):148. PMID: 34930452.5. Wilson PW, Meigs JB, Sullivan L, Fox CS, Nathan DM, D’Agostino RB Sr. Prediction of incident diabetes mellitus in middle-aged adults: the Framingham Offspring Study. Arch Intern Med. 2007; 167(10):1068–1074. PMID: 17533210.6. World Health Organization. Use of glycated haemoglobin (HbA1c) in the diagnosis of diabetes mellitus. Updated 2011. Accessed March 1, 2023. https://apps.who.int/iris/bitstream/handle/10665/70523/WHO_NMH_CHP_CPM_11.1_eng.pdf .7. Flack JM, Adekola B. Blood pressure and the new ACC/AHA hypertension guidelines. Trends Cardiovasc Med. 2020; 30(3):160–164. PMID: 31521481.8. Kim HL, Lee EM, Ahn SY, Kim KI, Kim HC, Kim JH, et al. The 2022 focused update of the 2018 Korean Hypertension Society guidelines for the management of hypertension. Clin Hypertens. 2023; 29(1):11. PMID: 36788612.9. Kim Y, Han BG. KoGES group. Cohort profile: the Korean Genome and Epidemiology Study (KoGES) consortium. Int J Epidemiol. 2017; 46(2):e20. PMID: 27085081.10. Collins GS, Mallett S, Omar O, Yu LM. Developing risk prediction models for type 2 diabetes: a systematic review of methodology and reporting. BMC Med. 2011; 9(1):103. PMID: 21902820.11. Mashayekhi M, Prescod F, Shah B, Dong L, Keshavjee K, Guergachi A. Evaluating the performance of the Framingham Diabetes Risk Scoring Model in Canadian electronic medical records. Can J Diabetes. 2015; 39(2):152–156. PMID: 25577729.12. Choi HS, Lee SW, Kim JT, Lee HK. The association between pulmonary functions and incident diabetes: longitudinal analysis from the Ansung cohort in Korea. Diabetes Metab J. 2020; 44(5):699–710. PMID: 32431104.13. Moon S, Jang JY, Kim Y, Oh CM. Development and validation of a new diabetes index for the risk classification of present and new-onset diabetes: multicohort study. Sci Rep. 2021; 11(1):15748. PMID: 34344964.14. Kim KN, Oh SY, Hong YC. Associations of serum calcium levels and dietary calcium intake with incident type 2 diabetes over 10 years: the Korean Genome and Epidemiology Study (KoGES). Diabetol Metab Syndr. 2018; 10(1):50. PMID: 29946367.15. Park S, Kim C, Wu X. Development and validation of an insulin resistance predicting model using a machine-learning approach in a population-based cohort in Korea. Diagnostics (Basel). 2022; 12(1):212. PMID: 35054379.16. Chien K, Cai T, Hsu H, Su T, Chang W, Chen M, et al. A prediction model for type 2 diabetes risk among Chinese people. Diabetologia. 2009; 52(3):443–450. PMID: 19057891.17. Li J, Bornstein SR, Landgraf R, Schwarz PE. Validation of a simple clinical diabetes prediction model in a middle-aged, white, German population. Arch Intern Med. 2007; 167(22):2528–2529.18. Lyssenko V, Jonsson A, Almgren P, Pulizzi N, Isomaa B, Tuomi T, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med. 2008; 359(21):2220–2232. PMID: 19020324.19. Nichols GA, Brown JB. Validating the Framingham Offspring Study equations for predicting incident diabetes mellitus. Am J Manag Care. 2008; 14(9):574–580. PMID: 18778172.20. Lindström J, Tuomilehto J. The diabetes risk score: a practical tool to predict type 2 diabetes risk. Diabetes Care. 2003; 26(3):725–731. PMID: 12610029.21. Sherwani SI, Khan HA, Ekhzaimy A, Masood A, Sakharkar MK. Significance of HbA1c test in diagnosis and prognosis of diabetic patients. Biomark Insights. 2016; 11:95–104. PMID: 27398023.22. Nathan DM, Turgeon H, Regan S. Relationship between glycated haemoglobin levels and mean glucose levels over time. Diabetologia. 2007; 50(11):2239–2244. PMID: 17851648.23. Stern MP, Williams K, Haffner SM. Identification of persons at high risk for type 2 diabetes mellitus: do we need the oral glucose tolerance test? Ann Intern Med. 2002; 136(8):575–581. PMID: 11955025.24. McNeely MJ, Boyko EJ, Leonetti DL, Kahn SE, Fujimoto WY. Comparison of a clinical model, the oral glucose tolerance test, and fasting glucose for prediction of type 2 diabetes risk in Japanese Americans. Diabetes Care. 2003; 26(3):758–763. PMID: 12610034.25. Shin J, Kim J, Lee C, Yoon JY, Kim S, Song S, et al. Development of various diabetes prediction models using machine learning techniques. Diabetes Metab J. 2022; 46(4):650–657. PMID: 35272434.26. Jeong YW, Jung Y, Jeong H, Huh JH, Sung KC, Shin JH, et al. Prediction model for hypertension and diabetes mellitus using Korean public health examination data (2002–2017). Diagnostics (Basel). 2022; 12(8):1967. PMID: 36010317.27. Jeon JY, Ko SH, Kwon HS, Kim NH, Kim JH, Kim CS, et al. Prevalence of diabetes and prediabetes according to fasting plasma glucose and HbA1c. Diabetes Metab J. 2013; 37(5):349–357. PMID: 24199164.28. Kaur G, Lakshmi PV, Rastogi A, Bhansali A, Jain S, Teerawattananon Y, et al. Diagnostic accuracy of tests for type 2 diabetes and prediabetes: a systematic review and meta-analysis. PLoS One. 2020; 15(11):e0242415. PMID: 33216783.29. Kashima S, Inoue K, Matsumoto M, Akimoto K. White blood cell count and C-reactive protein independently predicted incident diabetes: Yuport Medical Checkup Center Study. Endocr Res. 2019; 44(4):127–137. PMID: 30895902.30. Chen JY, Chen YH, Lee YC, Tsou MT. The association between white blood cell count and insulin resistance in community-dwelling middle-aged and older populations in Taiwan: a community-based cross-sectional study. Front Med (Lausanne). 2022; 9:813222. PMID: 35252251.31. Onur S, Niklowitz P, Jacobs G, Nöthlings U, Lieb W, Menke T, et al. Ubiquinol reduces gamma glutamyltransferase as a marker of oxidative stress in humans. BMC Res Notes. 2014; 7(1):427. PMID: 24996614.32. Onat A, Can G, Örnek E, Çiçek G, Ayhan E, Doğan Y. Serum γ-glutamyltransferase: independent predictor of risk of diabetes, hypertension, metabolic syndrome, and coronary disease. Obesity (Silver Spring). 2012; 20(4):842–848. PMID: 21633402.33. Lee JH, Lee HS, Lee YJ. Serum γ-glutamyltransferase as an independent predictor for incident type 2 diabetes in middle-aged and older adults: findings from the KoGES over 12 years of follow-up. Nutr Metab Cardiovasc Dis. 2020; 30(9):1484–1491. PMID: 32600956.34. Cho HC. The association between serum GGT concentration and diabetic peripheral polyneuropathy in type 2 diabetic patients. Korean Diabetes J. 2010; 34(2):111–118. PMID: 20548843.35. Valizadeh N, Mohammadi R, Mehdizadeh A, Motarjemizadeh Q, Khalkhali HR. Evaluation of serum gamma glutamyl transferase levels in diabetic patients with and without retinopathy. Shiraz E Med J. 2018; 19(7):e64073.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Validation of Risk Prediction Models for Atherosclerotic Cardiovascular Disease in a Prospective Korean Community-Based Cohort
- Framingham Equation Model Overestimates Risk of Ischemic Heart Disease in Korean Men and Women
- Exploration and Validation of the Performance of Hemoglobin A1c in Detecting Diabetes in CommunityDwellers With Hypertension
- Diagnostic Accuracy of Machine Learning Algorithms for Hepatitis A Antibody
- Assessment of Metabolic Syndrome Risk Based on Body Size Phenotype in Korean Adults: Analysis of Community-based Cohort Data