Cancer Res Treat.
2024 Jan;56(1):259-271. 10.4143/crt.2022.1600.
Gemcitabine Inhibits the Progression of Pancreatic Cancer by Restraining the WTAP/MYC Chain in an m6A-Dependent Manner
- Affiliations
-
- 1Department of General Surgery,The First Affiliated Hospital of Soochow University, Suzhou, China
- 2Department of Infectious Disease,The First Affiliated Hospital of Soochow University, Suzhou, China
- KMID: 2550341
- DOI: http://doi.org/10.4143/crt.2022.1600
Abstract
- Purpose
Pancreatic cancer (PC) is a common malignant tumor of the digestive system, and its 5-year survival rate is only 4%. N6-methyladenosine (m6A) RNA methylation is the most common post-transcriptional modification and dynamically regulates cancer development, while its role in PC treatment remains unclear.
Materials and Methods
We treated PC cells with gemcitabine and quantified the overall m6A level with m6A methylation quantification. Real-time quantitative reverse transcription polymerase chain reaction and Western blot analyses were used to detect expression changes of m6A regulators. We verified the m6A modification on the target genes through m6A-immunoprecipitation (IP), and further in vivo experiments and immunofluorescence (IF) assays were applied to verify regulation of gemcitabine on Wilms’ tumor 1–associated protein (WTAP) and MYC.
Results
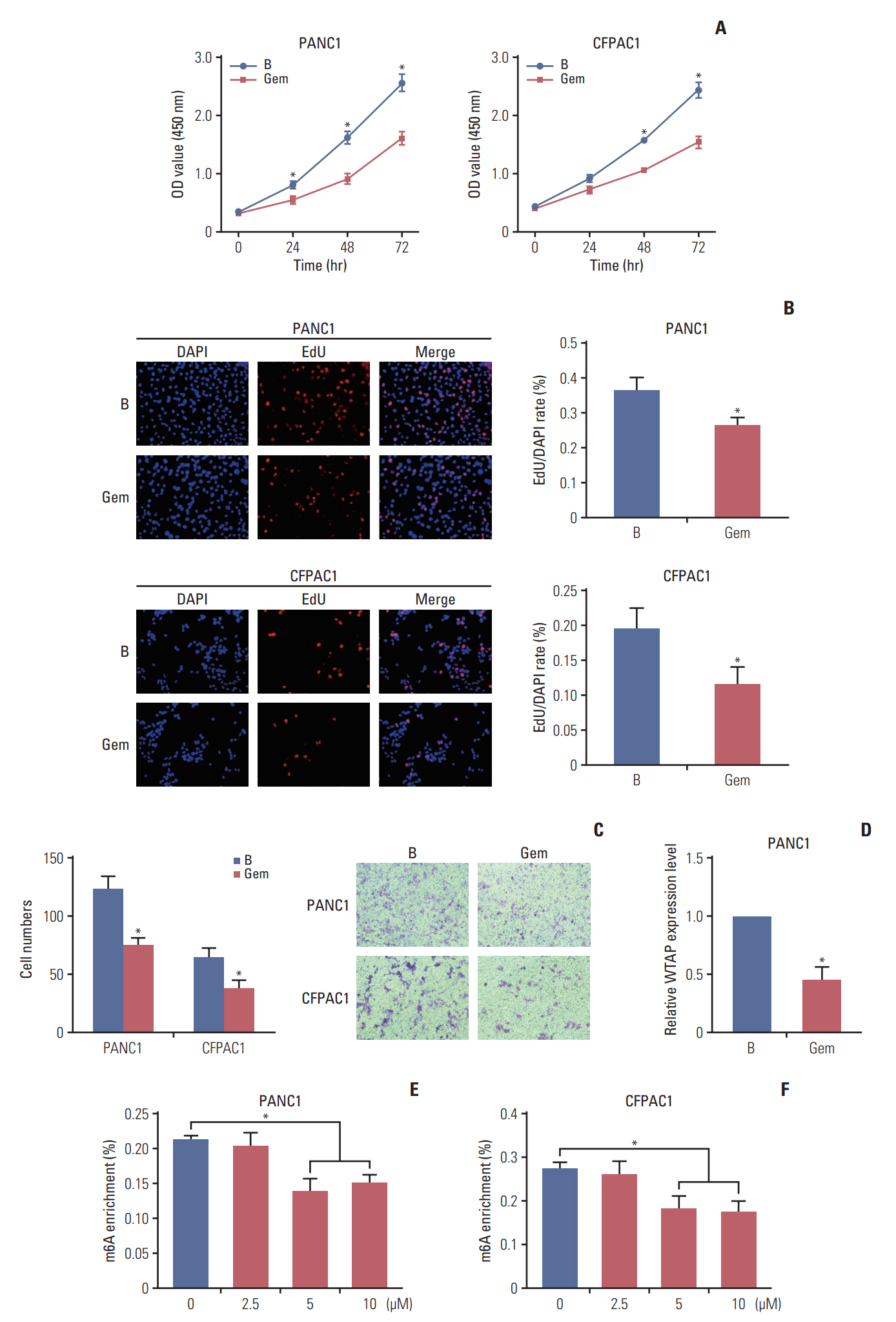
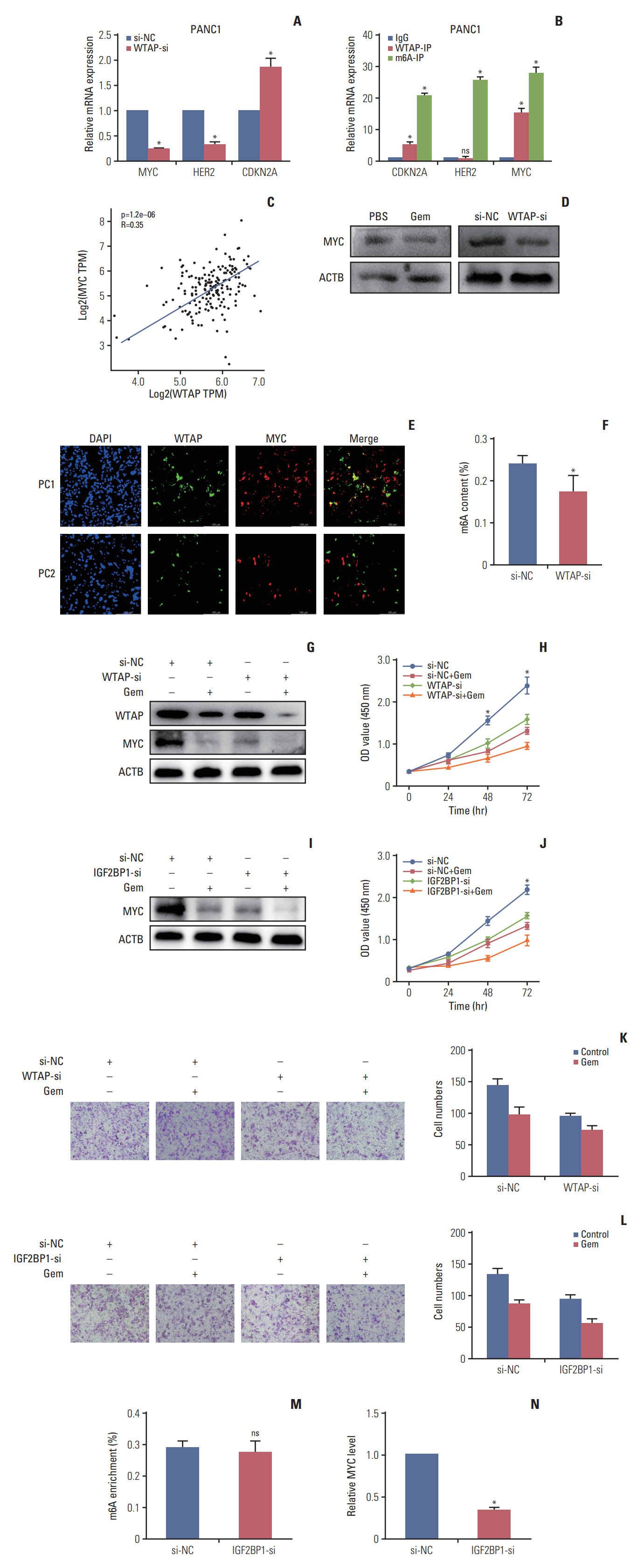
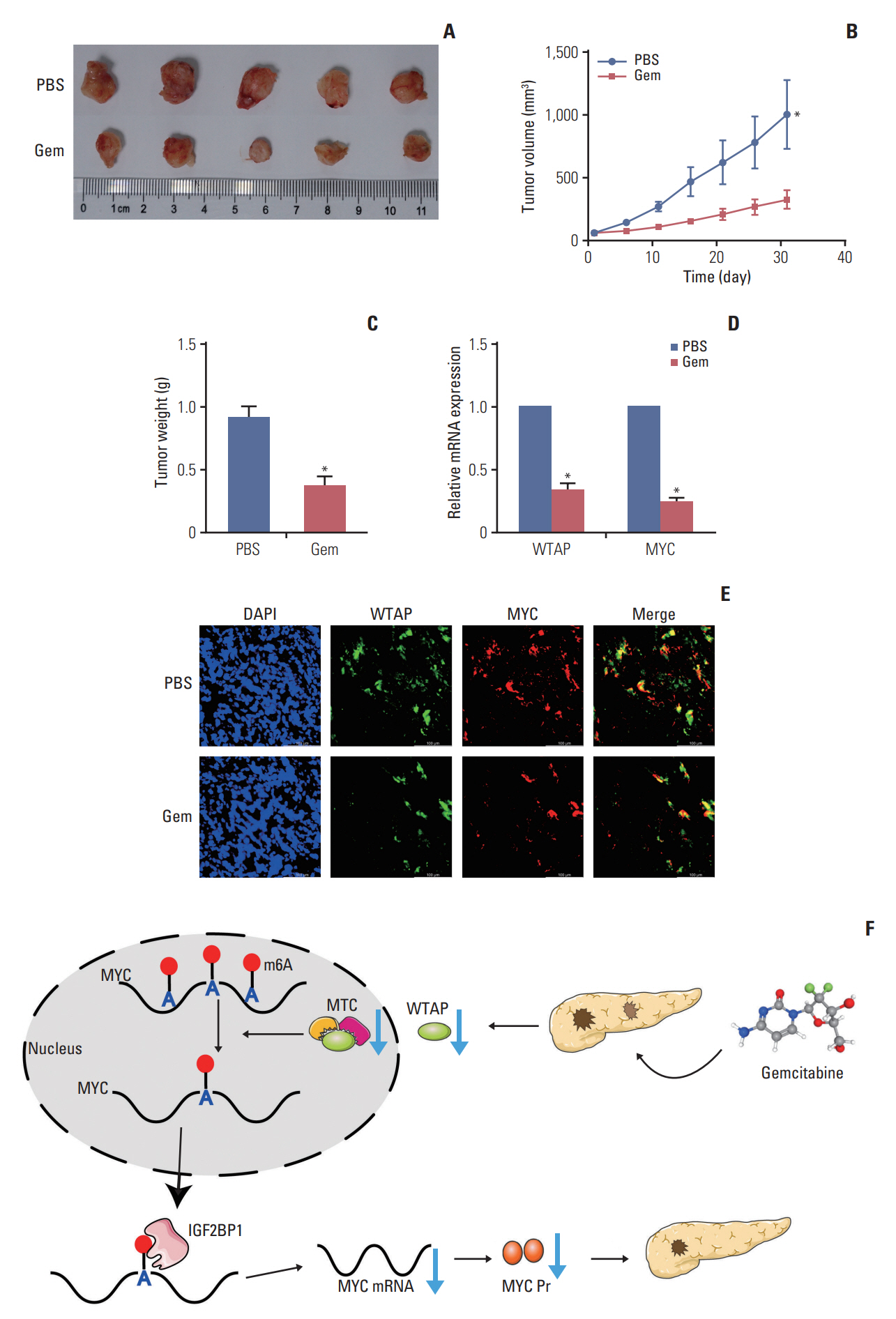
Gemcitabine inhibited the proliferation and migration of PC cells and reduced the overall level of m6A modification. Additionally, the expression of the “writer” WTAP was significantly downregulated after gemcitabine treatment. We knocked down WTAP in cells and found target gene MYC expression was significantly downregulated, m6A-IP also confirmed the m6A modification on MYC. Our experiments showed that m6A-MYC may be recognized by the “reader” IGF2BP1. In vivo experiments revealed gemcitabine inhibited the tumorigenic ability of PC cells. IF analysis also showed that gemcitabine inhibited the expression of WTAP and MYC, which displayed a significant trend of co-expression.
Conclusion
Our study confirmed that gemcitabine interferes with WTAP protein expression in PC, reduces m6A modification on MYC and RNA stability, thereby inhibiting the downstream pathway of MYC, and inhibits the progression of PC.
Keyword
Figure
Reference
-
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019; 69:7–34.2. Neoptolemos JP, Stocken DD, Friess H, Bassi C, Dunn JA, Hickey H, et al. A randomized trial of chemoradiotherapy and chemotherapy after resection of pancreatic cancer. N Engl J Med. 2004; 350:1200–10.3. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021; 71:7–33.4. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017; 169:1187–200.5. Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017; 18:31–42.6. Guo X, Li K, Jiang W, Hu Y, Xiao W, Huang Y, et al. RNA demethylase ALKBH5 prevents pancreatic cancer progression by posttranscriptional activation of PER1 in an m6A-YTHDF2-dependent manner. Mol Cancer. 2020; 19:91.7. Hu X, Peng WX, Zhou H, Jiang J, Zhou X, Huang D, et al. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 2020; 27:1782–94.8. Wang M, Liu J, Zhao Y, He R, Xu X, Guo X, et al. Upregulation of METTL14 mediates the elevation of PERP mRNA N6 adenosine methylation promoting the growth and metastasis of pancreatic cancer. Mol Cancer. 2020; 19:130.9. Taketo K, Konno M, Asai A, Koseki J, Toratani M, Satoh T, et al. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int J Oncol. 2018; 52:621–9.10. Hertel LW, Boder GB, Kroin JS, Rinzel SM, Poore GA, Todd GC, et al. Evaluation of the antitumor activity of gemcitabine (2’,2’-difluoro-2’-deoxycytidine). Cancer Res. 1990; 50:4417–22.11. Xu BQ, Fu ZG, Meng Y, Wu XQ, Wu B, Xu L, et al. Gemcitabine enhances cell invasion via activating HAb18G/CD147-EGFR-pSTAT3 signaling. Oncotarget. 2016; 7:62177–93.12. Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009; 458:762–5.13. Dong P, Maddali MV, Srimani JK, Thelot F, Nevins JR, Mathey-Prevot B, et al. Division of labour between Myc and G1 cyclins in cell cycle commitment and pace control. Nat Commun. 2014; 5:4750.14. Zhu S, Wang JZ, Chen D, He YT, Meng N, Chen M, et al. An oncopeptide regulates m6A recognition by the m6A reader IGF2BP1 and tumorigenesis. Nat Commun. 2020; 11:1685.15. Zhu P, He F, Hou Y, Tu G, Li Q, Jin T, et al. A novel hypoxic long noncoding RNA KB-1980E6.3 maintains breast cancer stem cell stemness via interacting with IGF2BP1 to facilitate c-Myc mRNA stability. Oncogene. 2021; 40:1609–27.16. Kleeff J, Reiser C, Hinz U, Bachmann J, Debus J, Jaeger D, et al. Surgery for recurrent pancreatic ductal adenocarcinoma. Ann Surg. 2007; 245:566–72.17. Raimondi S, Maisonneuve P, Lowenfels AB. Epidemiology of pancreatic cancer: an overview. Nat Rev Gastroenterol Hepatol. 2009; 6:699–708.18. Omura N, Goggins M. Epigenetics and epigenetic alterations in pancreatic cancer. Int J Clin Exp Pathol. 2009; 2:310–26.19. Huxley R, Ansary-Moghaddam A, Berrington de Gonzalez A, Barzi F, Woodward M. Type-II diabetes and pancreatic cancer: a meta-analysis of 36 studies. Br J Cancer. 2005; 92:2076–83.20. Duell EJ, Lucenteforte E, Olson SH, Bracci PM, Li D, Risch HA, et al. Pancreatitis and pancreatic cancer risk: a pooled analysis in the International Pancreatic Cancer Case-Control Consortium (PanC4). Ann Oncol. 2012; 23:2964–70.21. Ligorio M, Sil S, Malagon-Lopez J, Nieman LT, Misale S, Di Pilato M, et al. Stromal microenvironment shapes the intratumoral architecture of pancreatic cancer. Cell. 2019; 178:160–75e27.22. Zhu H, Li T, Du Y, Li M. Pancreatic cancer: challenges and opportunities. BMC Med. 2018; 16:214.23. Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer. 2020; 20:303–22.24. Zhou Z, Lv J, Yu H, Han J, Yang X, Feng D, et al. Mechanism of RNA modification N6-methyladenosine in human cancer. Mol Cancer. 2020; 19:104.25. Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019; 18:110.26. Yang X, Zhang S, He C, Xue P, Zhang L, He Z, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020; 19:46.27. Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020; 48:3816–31.28. Heinemann V, Quietzsch D, Gieseler F, Gonnermann M, Schonekas H, Rost A, et al. Randomized phase III trial of gemcitabine plus cisplatin compared with gemcitabine alone in advanced pancreatic cancer. J Clin Oncol. 2006; 24:3946–52.29. Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006; 16:253–64.30. Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008; 8:976–90.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Second Line Chemotherapy for Pancreatic Cancer
- Chemotherapy for Pancreatic Cancer
- α, γ-Mangostins Induce Autophagy and Show Synergistic Effect with Gemcitabine in Pancreatic Cancer Cell Lines
- A Phase II Study of Combination Chemotherapy with Gemcitabine, 5-fluorouracil, and Cisplatin for Advanced Pancreatic Cancer
- Long Term Complete Response of Unresectable Locally Advanced Pancreatic Cancer after CCRT and Gemcitabine Chemotherapy