Korean J Physiol Pharmacol.
2024 Jan;28(1):83-91. 10.4196/kjpp.2024.28.1.83.
ACY-241, a histone deacetylase 6 inhibitor, suppresses the epithelial–mesenchymal transition in lung cancer cells by downregulating hypoxia-inducible factor-1 alpha
- Affiliations
-
- 1College of Pharmacy, Keimyung University, Daegu 42601, Korea
- KMID: 2550291
- DOI: http://doi.org/10.4196/kjpp.2024.28.1.83
Abstract
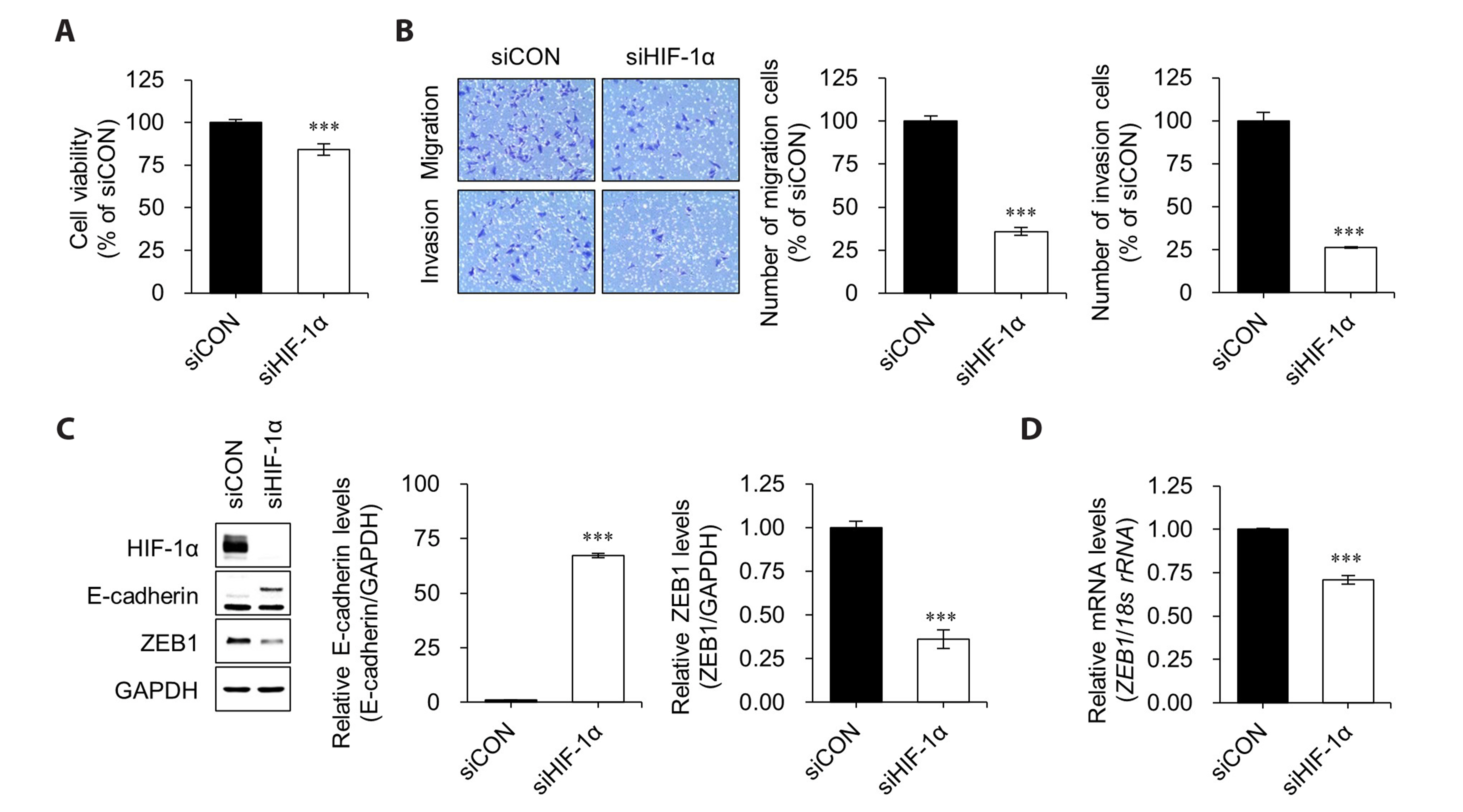
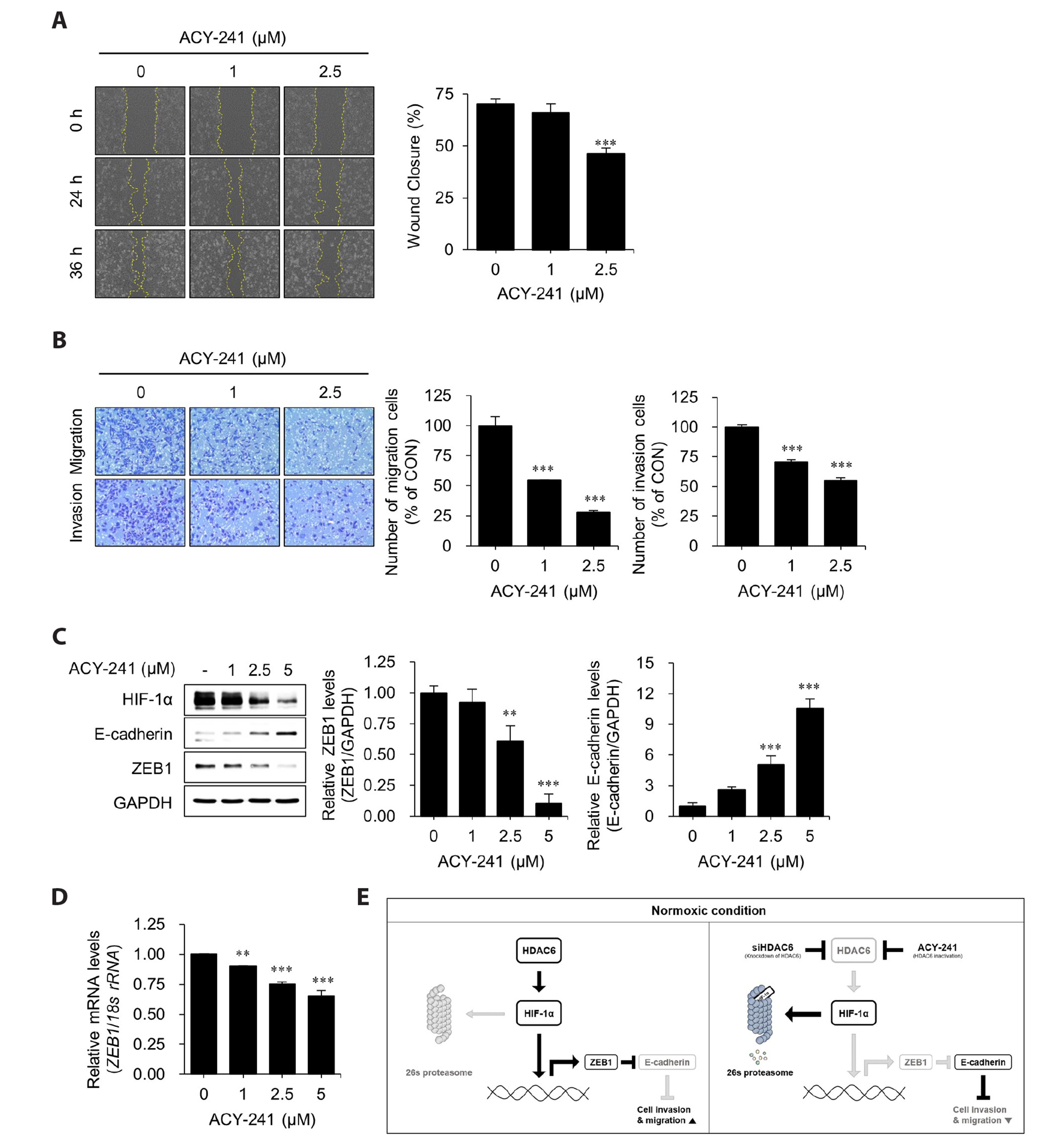
- Hypoxia-inducible factor-1 alpha (HIF-1α) is a transcription factor activated under hypoxic conditions, and it plays a crucial role in cellular stress regulation. While HIF-1α activity is essential in normal tissues, its presence in the tumor microenvironment represents a significant risk factor as it can induce angiogenesis and confer resistance to anti-cancer drugs, thereby contributing to poor prognoses. Typically, HIF-1α undergoes rapid degradation in normoxic conditions via oxygen-dependent degradation mechanisms. However, certain cancer cells can express HIF-1α even under normoxia. In this study, we observed an inclination toward increased normoxic HIF-1α expression in cancer cell lines exhibiting increased HDAC6 expression, which prompted the hypothesis that HDAC6 may modulate HIF-1α stability in normoxic conditions. To prove this hypothesis, several cancer cells with relatively higher HIF-1α levels under normoxic conditions were treated with ACY-241, a selective HDAC6 inhibitor, and small interfering RNAs for HDAC6 knockdown. Our data revealed a significant reduction in HIF-1α expression upon HDAC6 inhibition. Moreover, the downregulation of HIF-1α under normoxic conditions decreased zinc finger E-box-binding homeobox 1 expression and increased E-cadherin levels in lung cancer H1975 cells, consequently suppressing cell invasion and migration. ACY-241 treatment also demonstrated an inhibitory effect on cell invasion and migration by reducing HIF-1α level. This study confirms that HDAC6 knockdown and ACY-241 treatment effectively decrease HIF-1α expression under normoxia, thereby suppressing the epithelial– mesenchymal transition. These findings highlight the potential of selective HDAC6 inhibition as an innovative therapeutic strategy for lung cancer.
Keyword
Figure
Reference
-
1. Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. 2021; Lung cancer. Lancet. 398:535–554. DOI: 10.1016/S0140-6736(21)00312-3. PMID: 34273294.2. Heist RS, Sequist LV, Engelman JA. 2012; Genetic changes in squamous cell lung cancer: a review. J Thorac Oncol. 7:924–933. DOI: 10.1097/JTO.0b013e31824cc334. PMID: 22722794. PMCID: PMC3404741.3. Dong J, Li B, Lin D, Zhou Q, Huang D. 2019; Advances in targeted therapy and immunotherapy for non-small cell lung cancer based on accurate molecular typing. Front Pharmacol. 10:230. DOI: 10.3389/fphar.2019.00230. PMID: 30930778. PMCID: PMC6424010. PMID: a06463d539ec4ce7942b7834c48c2287.4. Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. 2009; An operational definition of epigenetics. Genes Dev. 23:781–783. DOI: 10.1101/gad.1787609. PMID: 19339683. PMCID: PMC3959995.5. Luo J, Huang Z, Wei W, Sun Y, Gong Y. 2023; Editorial: epigenetic regulation and non-histone post-translational modification in cancer. Front Genet. 14:1176174. DOI: 10.3389/fgene.2023.1176174. PMID: 37091796. PMCID: PMC10116044. PMID: 88d2c1c7ab0e4253b85f93110455a688.6. Delcuve GP, Khan DH, Davie JR. 2012; Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin Epigenetics. 4:5. DOI: 10.1186/1868-7083-4-5. PMID: 22414492. PMCID: PMC3320549. PMID: 3837566c859448f1ae5943186a7d0d81.7. Aldana-Masangkay GI, Sakamoto KM. 2011; The role of HDAC6 in cancer. J Biomed Biotechnol. 2011:875824. DOI: 10.1155/2011/875824. PMID: 21076528. PMCID: PMC2975074.8. Wang Z, Tang F, Hu P, Wang Y, Gong J, Sun S, Xie C. 2016; HDAC6 promotes cell proliferation and confers resistance to gefitinib in lung adenocarcinoma. Oncol Rep. 36:589–597. DOI: 10.3892/or.2016.4811. PMID: 27221381.9. Pulya S, Amin SA, Adhikari N, Biswas S, Jha T, Ghosh B. 2021; HDAC6 as privileged target in drug discovery: a perspective. Pharmacol Res. 163:105274. DOI: 10.1016/j.phrs.2020.105274. PMID: 33171304.10. Dong J, Zheng N, Wang X, Tang C, Yan P, Zhou HB, Huang J. 2018; A novel HDAC6 inhibitor exerts an anti-cancer effect by triggering cell cycle arrest and apoptosis in gastric cancer. Eur J Pharmacol. 828:67–79. DOI: 10.1016/j.ejphar.2018.03.026. PMID: 29563065.11. Li J, Yu M, Fu S, Liu D, Tan Y. 2022; Role of selective histone deacetylase 6 inhibitor ACY-1215 in cancer and other human diseases. Front Pharmacol. 13:907981. Erratum in: Front Pharmacol. 2022;13: 1117936. DOI: 10.3389/fphar.2022.907981. PMID: 35652048. PMCID: PMC9149003. PMID: d5a8624566a44df2aec9a2d8779e4720.12. Park SJ, Joo SH, Lee N, Jang WJ, Seo JH, Jeong CH. 2021; ACY-241, an HDAC6 inhibitor, overcomes erlotinib resistance in human pancreatic cancer cells by inducing autophagy. Arch Pharm Res. 44:1062–1075. DOI: 10.1007/s12272-021-01359-x. PMID: 34761352.13. Zhang XH, Kang HQ, Tao YY, Li YH, Zhao JR, Ma LY, Liu HM. Ya-Gao. 2021; Identification of novel 1,3-diaryl-1,2,4-triazole-capped histone deacetylase 6 inhibitors with potential anti-gastric cancer activity. Eur J Med Chem. 218:113392. DOI: 10.1016/j.ejmech.2021.113392. PMID: 33831778.14. Yang MH, Wu KJ. 2008; TWIST activation by hypoxia inducible factor-1 (HIF-1): implications in metastasis and development. Cell Cycle. 7:2090–2096. DOI: 10.4161/cc.7.14.6324. PMID: 18635960.15. Jun JC, Rathore A, Younas H, Gilkes D, Polotsky VY. 2017; Hypoxia-inducible factors and cancer. Curr Sleep Med Rep. 3:1–10. DOI: 10.1007/s40675-017-0062-7. PMID: 28944164. PMCID: PMC5607450.16. Kurtipek E, Koçak N, Esme H, Düzgün N, Akin SE, Ünlü Y, Bekçi TT. 2016; The role of HIF-1 pathway in non-small-cell lung cancer. Eur Respir J. 48:PA2855. DOI: 10.1183/13993003.congress-2016.PA2855.17. Zhu J, Huang Z, Zhang M, Wang W, Liang H, Zeng J, Wu K, Wang X, Hsieh JT, Guo P, Fan J. 2018; HIF-1α promotes ZEB1 expression and EMT in a human bladder cancer lung metastasis animal model. Oncol Lett. 15:3482–3489. Erratum in: Oncol Lett. 2021;22:599. DOI: 10.3892/ol.2021.12860. PMID: 34188701. PMCID: PMC8228379.18. Stroka DM, Burkhardt T, Desbaillets I, Wenger RH, Neil DA, Bauer C, Gassmann M, Candinas D. 2001; HIF-1 is expressed in normoxic tissue and displays an organ-specific regulation under systemic hypoxia. FASEB J. 15:2445–2453. DOI: 10.1096/fj.01-0125com. PMID: 11689469.19. Cao Y, Eble JM, Moon E, Yuan H, Weitzel DH, Landon CD, Nien CY, Hanna G, Rich JN, Provenzale JM, Dewhirst MW. 2013; Tumor cells upregulate normoxic HIF-1α in response to doxorubicin. Cancer Res. 73:6230–6242. DOI: 10.1158/0008-5472.CAN-12-1345. PMID: 23959856. PMCID: PMC3800255.20. Ryu HW, Won HR, Lee DH, Kwon SH. 2017; HDAC6 regulates sensitivity to cell death in response to stress and post-stress recovery. Cell Stress Chaperones. 22:253–261. DOI: 10.1007/s12192-017-0763-3. PMID: 28116619. PMCID: PMC5352599.21. Livak KJ, Schmittgen TD. 2001; Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. DOI: 10.1006/meth.2001.1262. PMID: 11846609.22. Zhang Q, Zhang ZF, Rao JY, Sato JD, Brown J, Messadi DV, Le AD. 2004; Treatment with siRNA and antisense oligonucleotides targeted to HIF-1alpha induced apoptosis in human tongue squamous cell carcinomas. Int J Cancer. 111:849–857. DOI: 10.1002/ijc.20334. PMID: 15300796.23. Masoud GN, Li W. 2015; HIF-1α pathway: role, regulation and intervention for cancer therapy. Acta Pharm Sin B. 5:378–389. DOI: 10.1016/j.apsb.2015.05.007. PMID: 26579469. PMCID: PMC4629436. PMID: 0f5f4982b0944ba585e72b39876cfd33.24. Sánchez-Tilló E, Lázaro A, Torrent R, Cuatrecasas M, Vaquero EC, Castells A, Engel P, Postigo A. 2010; ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene. 29:3490–3500. DOI: 10.1038/onc.2010.102. PMID: 20418909.25. Haase VH. 2009; The VHL tumor suppressor: master regulator of HIF. Curr Pharm Des. 15:3895–3903. DOI: 10.2174/138161209789649394. PMID: 19671042. PMCID: PMC3622710.26. Li T, Mao C, Wang X, Shi Y, Tao Y. 2020; Epigenetic crosstalk between hypoxia and tumor driven by HIF regulation. J Exp Clin Cancer Res. 39:224. DOI: 10.1186/s13046-020-01733-5. PMID: 33109235. PMCID: PMC7592369. PMID: 3dc64a595d5a44a6be5538d4e2133c8f.27. Liang D, Kong X, Sang N. 2006; Effects of histone deacetylase inhibitors on HIF-1. Cell Cycle. 5:2430–2435. DOI: 10.4161/cc.5.21.3409. PMID: 17102633. PMCID: PMC4505804.28. Yoo YG, Kong G, Lee MO. 2006; Metastasis-associated protein 1 enhances stability of hypoxia-inducible factor-1alpha protein by recruiting histone deacetylase 1. EMBO J. 25:1231–1241. DOI: 10.1038/sj.emboj.7601025. PMID: 16511565. PMCID: PMC1422150.29. Seo HW, Kim EJ, Na H, Lee MO. 2009; Transcriptional activation of hypoxia-inducible factor-1alpha by HDAC4 and HDAC5 involves differential recruitment of p300 and FIH-1. FEBS Lett. 583:55–60. DOI: 10.1016/j.febslet.2008.11.044. PMID: 19071119.30. Kong X, Lin Z, Liang D, Fath D, Sang N, Caro J. 2006; Histone deacetylase inhibitors induce VHL and ubiquitin-independent proteasomal degradation of hypoxia-inducible factor 1alpha. Mol Cell Biol. 26:2019–2028. DOI: 10.1128/MCB.26.6.2019-2028.2006. PMID: 16507982. PMCID: PMC1430285.31. Geng H, Liu Q, Xue C, David LL, Beer TM, Thomas GV, Dai MS, Qian DZ. 2012; HIF1α protein stability is increased by acetylation at lysine 709. J Biol Chem. 287:35496–35505. DOI: 10.1074/jbc.M112.400697. PMID: 22908229. PMCID: PMC3471753.32. Saito S, Zhuang Y, Shan B, Danchuk S, Luo F, Korfei M, Guenther A, Lasky JA. 2017; Tubastatin ameliorates pulmonary fibrosis by targeting the TGFβ-PI3K-Akt pathway. PLoS One. 12:e0186615. DOI: 10.1371/journal.pone.0186615. PMID: 29045477. PMCID: PMC5646855. PMID: e3513edce61f4cd49294030347401e17.33. Schoepflin ZR, Shapiro IM, Risbud MV. 2016; Class I and IIa HDACs mediate HIF-1α stability through PHD2-dependent mechanism, while HDAC6, a Class IIb member, promotes HIF-1α transcriptional activity in nucleus pulposus cells of the intervertebral disc. J Bone Miner Res. 31:1287–1299. DOI: 10.1002/jbmr.2787. PMID: 26765925. PMCID: PMC4891304.34. McLeod AB, Stice JP, Wardell SE, Alley HM, Chang CY, McDonnell DP. 2018; Validation of histone deacetylase 3 as a therapeutic target in castration-resistant prostate cancer. Prostate. 78:266–277. DOI: 10.1002/pros.23467. PMID: 29243324.35. Yang L, Chang Y, Cao P. 2018; CCR7 preservation via histone deacetylase inhibition promotes epithelial-mesenchymal transition of hepatocellular carcinoma cells. Exp Cell Res. 371:231–237. DOI: 10.1016/j.yexcr.2018.08.015. PMID: 30107147.36. Wang J, Xu MQ, Jiang XL, Mei XY, Liu XG. 2018; Histone deacetylase inhibitor SAHA-induced epithelial-mesenchymal transition by upregulating Slug in lung cancer cells. Anticancer Drugs. 29:80–88. DOI: 10.1097/CAD.0000000000000573. PMID: 29176396.37. Wawruszak A, Kalafut J, Okon E, Czapinski J, Halasa M, Przybyszewska A, Miziak P, Okla K, Rivero-Muller A, Stepulak A. 2019; Histone deacetylase inhibitors and phenotypical transformation of cancer cells. Cancers (Basel). 11:148. DOI: 10.3390/cancers11020148. PMID: 30691229. PMCID: PMC6406474. PMID: 542b51f83d824eb3b05c7dac0f82a30c.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Effects of Histone Deacetylase Inhibitor (Valproic Acid) on the Expression of Hypoxia-inducible Factor-1 Alpha in Human Retinal Müller Cells
- Wheatgrass extract inhibits hypoxia-inducible factor-1-mediated epithelial-mesenchymal transition in A549 cells
- The role of hypoxia on the acquisition of epithelial-mesenchymal transition and cancer stemness: a possible link to epigenetic regulation
- Trefoil Factor 1 Suppresses Epithelial-mesenchymal Transition through Inhibition of TGF-beta Signaling in Gastric Cancer Cells
- Hypoxia Induces Epithelial-Mesenchymal Transition in Follicular Thyroid Cancer: Involvement of Regulation of Twist by Hypoxia Inducible Factor-1alpha