Korean J Physiol Pharmacol.
2024 Jan;28(1):59-72. 10.4196/kjpp.2024.28.1.59.
Mechanism of Wenshen Xuanbi Decoction in the treatment of osteoarthritis based on network pharmacology and experimental verification
- Affiliations
-
- 1Department of Orthopedics, Affiliated Hospital of Nanjing University of Chinese Medicine, Nanjing 210029, Jiangsu, China
- 2Department of Orthopedics, Jiangsu Provincial Hospital of Chinese Medicine, Nanjing 210029, Jiangsu, China
- KMID: 2550289
- DOI: http://doi.org/10.4196/kjpp.2024.28.1.59
Abstract
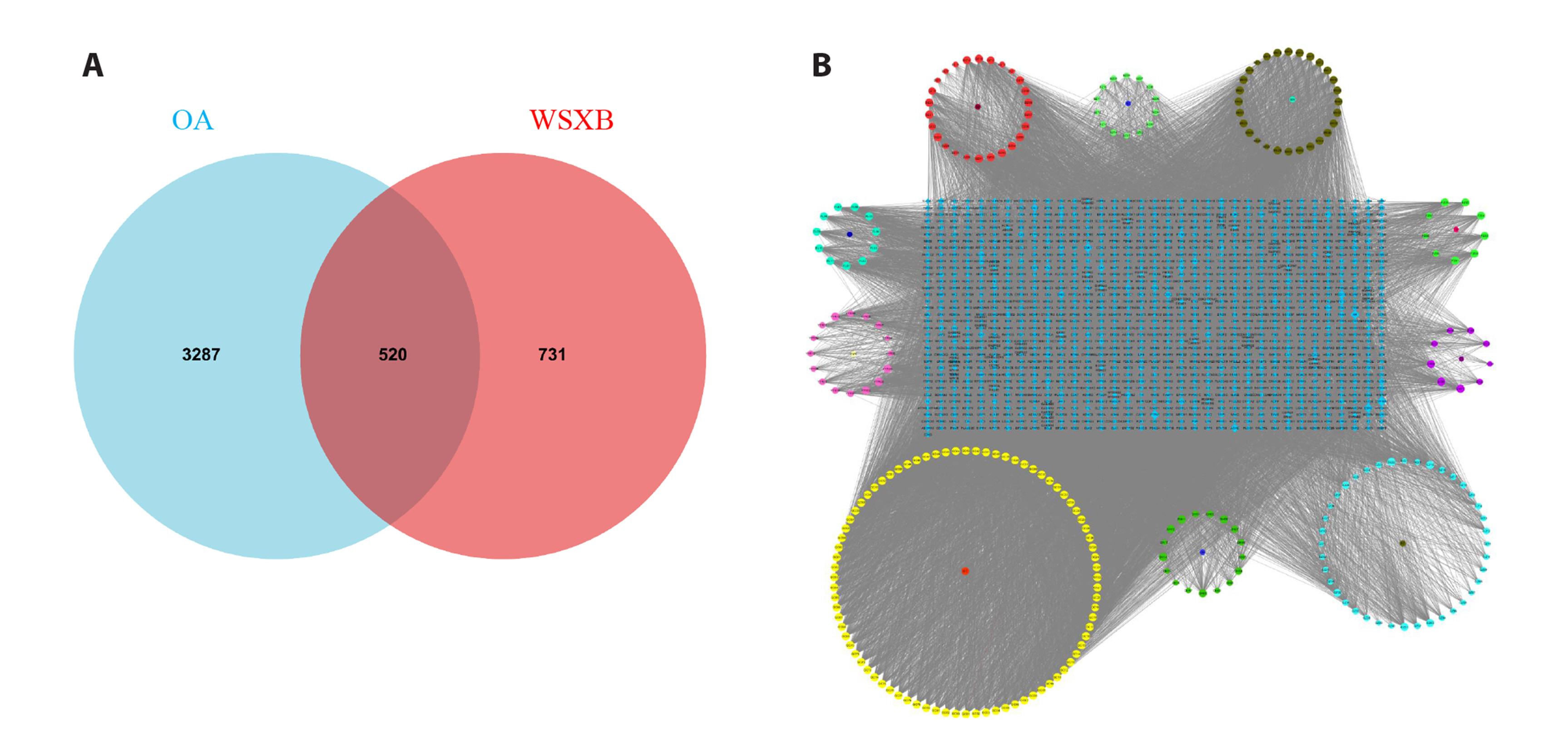
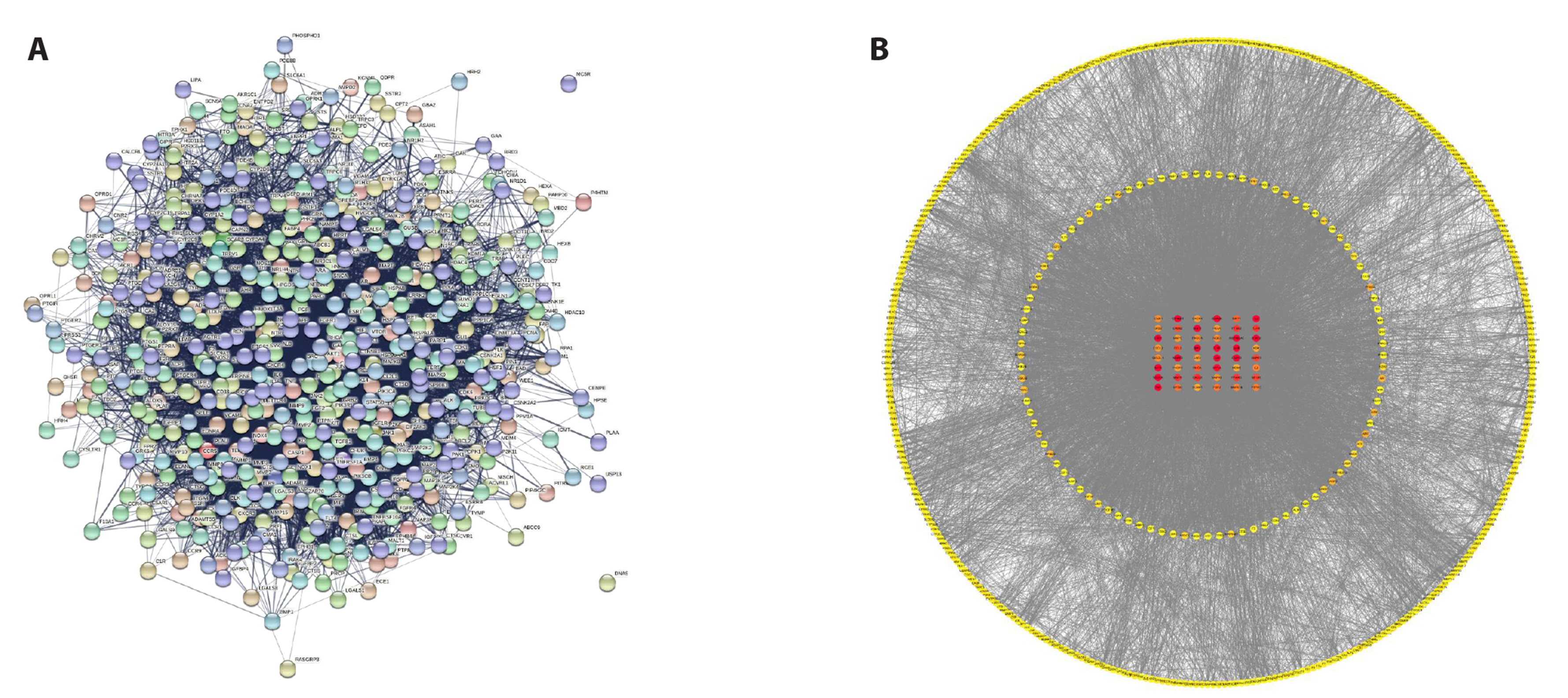
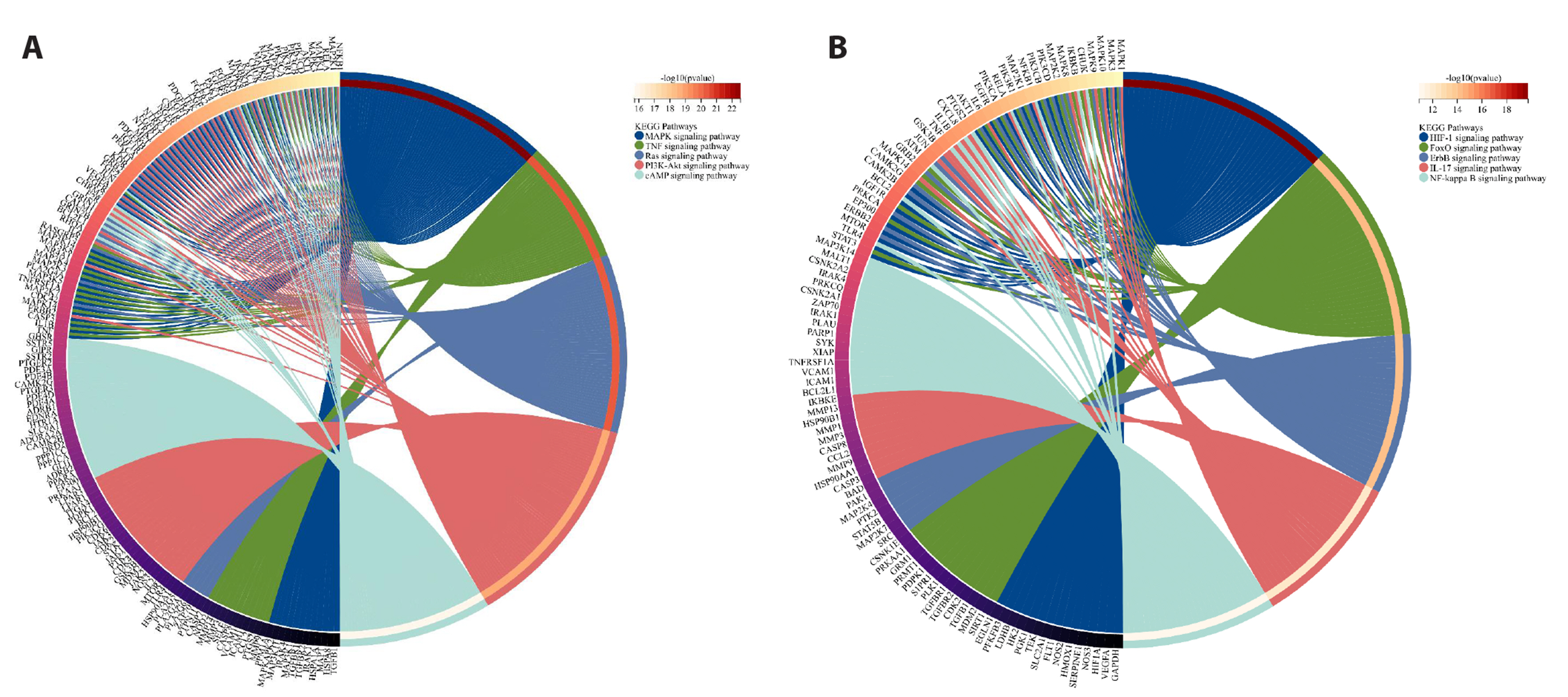
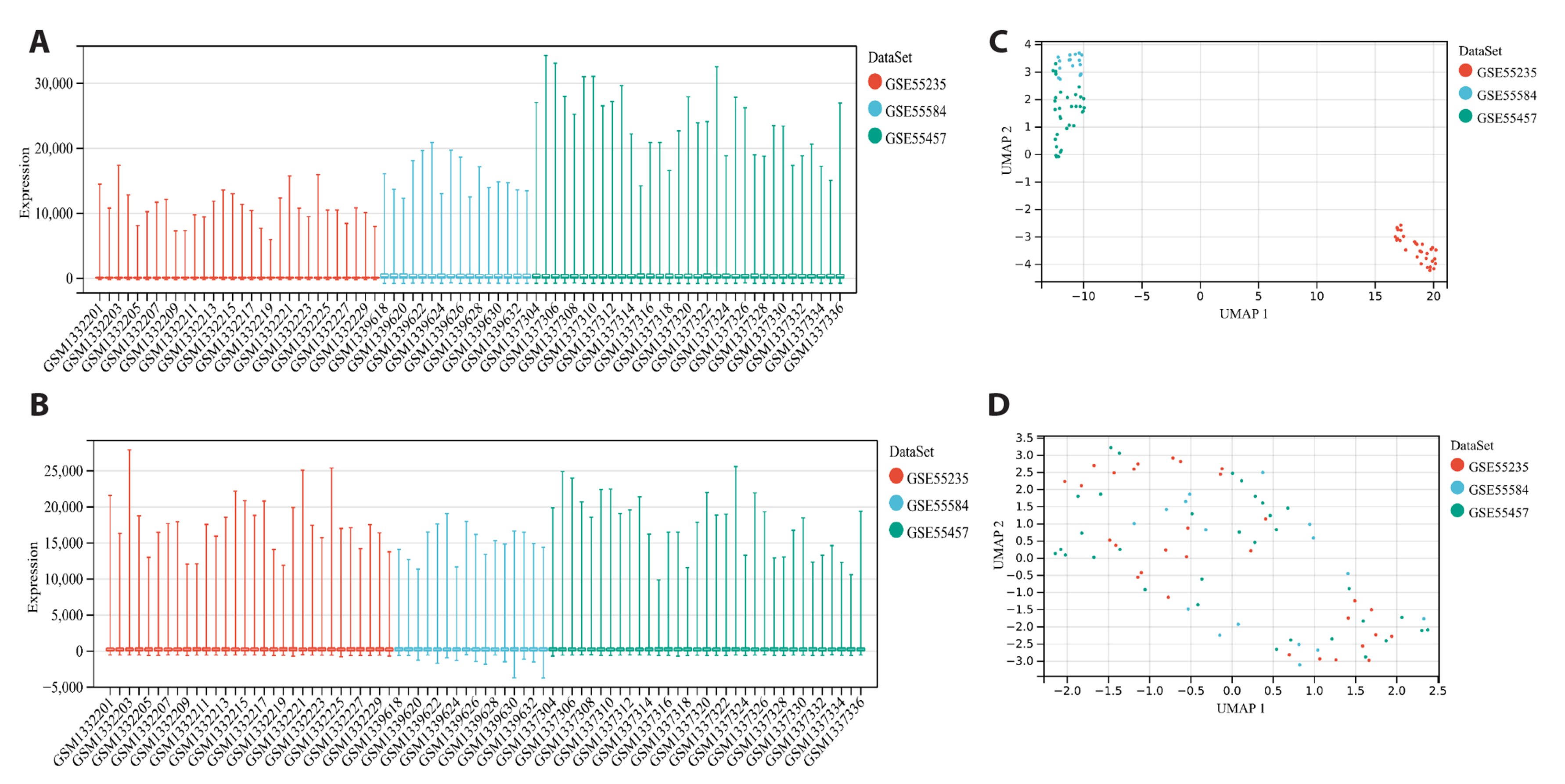
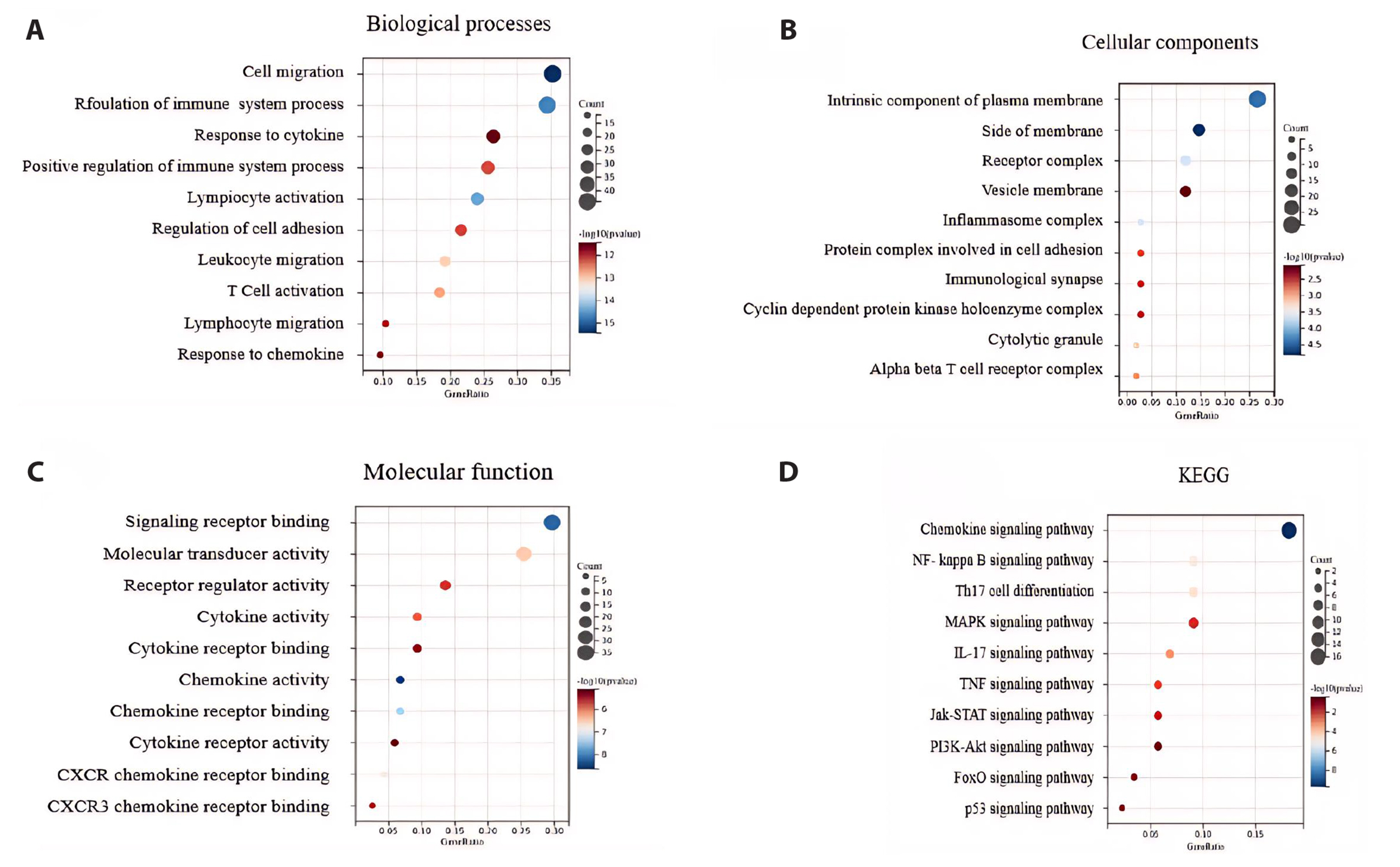
- To investigate the mechanism of Wenshen Xuanbi Decoction (WSXB) in treating osteoarthritis (OA) via network pharmacology, bioinformatics analysis, and experimental verification. The active components and prediction targets of WSXB were obtained from the TCMSP database and Swiss Target Prediction website, respectively. OA-related genes were retrieved from GeneCards and OMIM databases. Protein-protein interaction and functional enrichment analyses were performed, resulting in the construction of the Herb-Component-Target network. In addition, differential genes of OA were obtained from the GEO database to verify the potential mechanism of WSXB in OA treatment. Subsequently, potential active components were subjected to molecular verification with the hub targets. Finally, we selected the most crucial hub targets and pathways for experimental verification in vitro. The active components in the study included quercetin, linolenic acid, methyl linoleate, isobergapten, and beta-sitosterol. AKT1, tumor necrosis factor (TNF), interleukin (IL)-6, GAPDH, and CTNNB1 were identified as the most crucial hub targets. Molecular docking revealed that the active components and hub targets exhibited strong binding energy. Experimental verification demonstrated that the mRNA and protein expression levels of IL-6, IL-17, and TNF in the WSXB group were lower than those in the KOA group (p < 0.05). WSXB exhibits a chondroprotective effect on OA and delays disease progression. The mechanism is potentially related to the suppression of IL-17 and TNF signaling pathways and the down-regulation of IL-6.
Figure
Reference
-
1. McHugh D, Gil J. 2018; Senescence and aging: causes, consequences, and therapeutic avenues. J Cell Biol. 217:65–77. DOI: 10.1083/jcb.201708092. PMID: 29114066. PMCID: PMC5748990.2. Prieto-Alhambra D, Judge A, Javaid MK, Cooper C, Diez-Perez A, Arden NK. 2014; Incidence and risk factors for clinically diagnosed knee, hip and hand osteoarthritis: influences of age, gender and osteoarthritis affecting other joints. Ann Rheum Dis. 73:1659–1664. DOI: 10.1136/annrheumdis-2013-203355. PMID: 23744977. PMCID: PMC3875433.3. Chen FP, Chang CM, Hwang SJ, Chen YC, Chen FJ. 2014; Chinese herbal prescriptions for osteoarthritis in Taiwan: analysis of National Health Insurance dataset. BMC Complement Altern Med. 14:91. DOI: 10.1186/1472-6882-14-91. PMID: 24606767. PMCID: PMC3973832.4. Salvo F, Fourrier-Réglat A, Bazin F, Robinson P, Riera-Guardia N, Haag M, Caputi AP, Moore N, Sturkenboom MC, Pariente A. Investigators of Safety of Non-Steroidal Anti-Inflammatory Drugs: SOS Project. 2011; Cardiovascular and gastrointestinal safety of NSAIDs: a systematic review of meta-analyses of randomized clinical trials. Clin Pharmacol Ther. 89:855–866. DOI: 10.1038/clpt.2011.45. PMID: 21471964.5. Mei ZG, Cheng CG, Zheng JF. 2011; Observations on curative effect of high-frequency electric sparkle and point-injection therapy on knee osteoarthritis. J Tradit Chin Med. 31:311–315. DOI: 10.1016/S0254-6272(12)60010-7. PMID: 22462237.6. Liu W, Wu YH, Liu XY, Xue B, Shen W, Yang K. 2013; Metabolic regulatory and anti-oxidative effects of modified Bushen Huoxue decoction on experimental rabbit model of osteoarthritis. Chin J Integr Med. 19:459–463. DOI: 10.1007/s11655-011-0727-x. PMID: 22528754.7. Wu JJ, Guo ZZ, Zhu YF, Huang ZJ, Gong X, Li YH, Son WJ, Li XY, Lou YM, Zhu LJ, Lu LL, Liu ZQ, Liu L. 2018; A systematic review of pharmacokinetic studies on herbal drug Fuzi: implications for Fuzi as personalized medicine. Phytomedicine. 44:187–203. DOI: 10.1016/j.phymed.2018.03.001. PMID: 29526584.8. Yoo SR, Kim Y, Lee MY, Kim OS, Seo CS, Shin HK, Jeong SJ. 2016; Gyeji-tang water extract exerts anti-inflammatory activity through inhibition of ERK and NF-κB pathways in lipopolysaccharide-stimulated RAW 264.7 cells. BMC Complement Altern Med. 16:390. DOI: 10.1186/s12906-016-1366-8. PMID: 27733198. PMCID: PMC5062814.9. Yang M, Ji X, Zuo Z. 2018; Relationships between the toxicities of Radix Aconiti Lateralis Preparata (Fuzi) and the toxicokinetics of its main diester-diterpenoid alkaloids. Toxins (Basel). 10:391. DOI: 10.3390/toxins10100391. PMID: 30261585. PMCID: PMC6215299. PMID: fe63b130d8c24ac38090e6803a6f36e7.10. Kim MC, Kim DS, Kim SJ, Park J, Kim HL, Kim SY, Ahn KS, Jang HJ, Lee SG, Lee KM, Hong SH, Um JY. 2012; Eucommiae cortex inhibits TNF-α and IL-6 through the suppression of caspase-1 in lipopolysaccharide-stimulated mouse peritoneal macrophages. Am J Chin Med. 40:135–149. DOI: 10.1142/S0192415X12500115. PMID: 22298454.11. Feng C, Zhao M, Jiang L, Hu Z, Fan X. 2021; Mechanism of Modified Danggui Sini Decoction for knee osteoarthritis based on network pharmacology and molecular docking. Evid Based Complement Alternat Med. 2021:6680637. DOI: 10.1155/2021/6680637. PMID: 33628311. PMCID: PMC7895562.12. Xu Y, Li H, He X, Huang Y, Wang S, Wang L, Fu C, Ye H, Li X, Asakawa T. 2021; Identification of the key role of NF-κB signaling pathway in the treatment of osteoarthritis with Bushen Zhuangjin Decoction, a verification based on network pharmacology approach. Front Pharmacol. 12:637273. DOI: 10.3389/fphar.2021.637273. PMID: 33912052. PMCID: PMC8072665. PMID: c9433876dca844fda9d97d694e176751.13. Zhang X, Gao R, Zhou Z, Sun J, Tang X, Li J, Zhou X, Shen T. 2022; Uncovering the mechanism of Huanglian-Wuzhuyu herb pair in treating nonalcoholic steatohepatitis based on network pharmacology and experimental validation. J Ethnopharmacol. 296:115405. DOI: 10.1016/j.jep.2022.115405. PMID: 35644437.14. Ye J, Li L, Hu Z. 2021; Exploring the molecular mechanism of action of Yinchen Wuling powder for the treatment of hyperlipidemia, using network pharmacology, molecular docking, and molecular dynamics simulation. Biomed Res Int. 2021:9965906. DOI: 10.1155/2021/9965906. PMID: 34746316. PMCID: PMC8568510.15. Chen D, Shen J, Zhao W, Wang T, Han L, Hamilton JL, Im HJ. 2017; Osteoarthritis: toward a comprehensive understanding of pathological mechanism. Bone Res. 5:16044. DOI: 10.1038/boneres.2016.44. PMID: 28149655. PMCID: PMC5240031.16. Ru J, Li P, Wang J, Zhou W, Li B, Huang C, Li P, Guo Z, Tao W, Yang Y, Xu X, Li Y, Wang Y, Yang L. 2014; TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 6:13. DOI: 10.1186/1758-2946-6-13. PMID: 24735618. PMCID: PMC4001360.17. Li J, Zhao P, Li Y, Tian Y, Wang Y. 2015; Systems pharmacology-based dissection of mechanisms of Chinese medicinal formula Bufei Yishen as an effective treatment for chronic obstructive pulmonary disease. Sci Rep. 5:15290. DOI: 10.1038/srep15290. PMID: 26469778. PMCID: PMC4606809.18. Xu X, Zhang W, Huang C, Li Y, Yu H, Wang Y, Duan J, Ling Y. 2012; A novel chemometric method for the prediction of human oral bioavailability. Int J Mol Sci. 13:6964–6982. DOI: 10.3390/ijms13066964. PMID: 22837674. PMCID: PMC3397506. PMID: 5d3e3f3045ce4bd1b445e1abbd7786be.19. Cincilla G, Thormann M, Pons M. 2010; Structuring chemical space: similarity-based characterization of the PubChem database. Mol Inform. 29:37–49. DOI: 10.1002/minf.200900015. PMID: 27463847.20. Gfeller D, Grosdidier A, Wirth M, Daina A, Michielin O, Zoete V. 2014; SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res. 42:W32–W38. DOI: 10.1093/nar/gku293. PMID: 24792161. PMCID: PMC4086140.21. Lin Y, Hu Z. 2021; Bioinformatics analysis of candidate genes involved in ethanol-induced microtia pathogenesis based on a human genome database: GeneCards. Int J Pediatr Otorhinolaryngol. 142:110595. DOI: 10.1016/j.ijporl.2020.110595. PMID: 33418206.22. van Triest HJ, Chen D, Ji X, Qi S, Li-Ling J. 2011; PhenOMIM: an OMIM-based secondary database purported for phenotypic comparison. Annu Int Conf IEEE Eng Med Biol Soc. 2011:3589–3592. DOI: 10.1109/IEMBS.2011.6090600. PMID: 22255115.23. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. 2019; STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47:D607–D613. DOI: 10.1093/nar/gky1131. PMID: 30476243. PMCID: PMC6323986.24. Reimand J, Isserlin R, Voisin V, Kucera M, Tannus-Lopes C, Rostamianfar A, Wadi L, Meyer M, Wong J, Xu C, Merico D, Bader GD. 2019; Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc. 14:482–517. DOI: 10.1038/s41596-018-0103-9. PMID: 30664679. PMCID: PMC6607905.25. Kanehisa M, Goto S. 2000; KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30. DOI: 10.1093/nar/28.1.27. PMID: 10592173. PMCID: PMC102409.26. Sherman BT, Huang da W, Tan Q, Guo Y, Bour S, Liu D, Stephens R, Baseler MW, Lane HC, Lempicki RA. 2007; DAVID Knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics. 8:426. DOI: 10.1186/1471-2105-8-426. PMID: 17980028. PMCID: PMC2186358. PMID: 14dc8b36e8434399b030757c7f5ac300.27. Liu Z, Li Y, Han L, Li J, Liu J, Zhao Z, Nie W, Liu Y, Wang R. 2015; PDB-wide collection of binding data: current status of the PDBbind database. Bioinformatics. 31:405–412. DOI: 10.1093/bioinformatics/btu626. PMID: 25301850.28. Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, Zaslavsky L, Zhang J, Bolton EE. 2021; PubChem in 2021: new data content and improved web interfaces. Nucleic. Acids Res. 49:D1388–D1395. DOI: 10.1093/nar/gkaa971. PMID: 33151290. PMCID: PMC7778930.29. Seeliger D, de Groot BL. 2010; Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des. 24:417–422. DOI: 10.1007/s10822-010-9352-6. PMID: 20401516. PMCID: PMC2881210.30. Mooers BHM. 2020; Shortcuts for faster image creation in PyMOL. Protein Sci. 29:268–276. DOI: 10.1002/pro.3781. PMID: 31710740. PMCID: PMC6933860.31. Toro-Domínguez D, Martorell-Marugán J, López-Domínguez R, García-Moreno A, González-Rumayor V, Alarcón-Riquelme ME, Carmona-Sáez P. 2019; ImaGEO: integrative gene expression meta-analysis from GEO database. Bioinformatics. 35:880–882. DOI: 10.1093/bioinformatics/bty721. PMID: 30137226.32. Taminau J, Meganck S, Lazar C, Steenhoff D, Coletta A, Molter C, Duque R, de Schaetzen V, Weiss Solís DY, Bersini H, Nowé A. 2012; Unlocking the potential of publicly available microarray data using inSilicoDb and inSilicoMerging R/Bioconductor packages. BMC Bioinformatics. 13:335. DOI: 10.1186/1471-2105-13-335. PMID: 23259851. PMCID: PMC3568420. PMID: 506321dc462b4db0a844505b4bdb8bdb.33. Huang Y, Yang DD, Li XY, Fang DL, Zhou WJ. 2021; ZBP1 is a significant pyroptosis regulator for systemic lupus erythematosus. Ann Transl Med. 9:1773. DOI: 10.21037/atm-21-6193. PMID: 35071467. PMCID: PMC8756241.34. Fan Y, Xia T, Shen J. 2021; The study of protective efficacy of Wenshenxuanbi Decoction on chondrocyte of knee osteoarthritis in rats. Chin J Med & Orth. 6:5–8.35. Ganesan R, Doss HM, Rasool M. 2016; Majoon ushba, a polyherbal compound, suppresses pro-inflammatory mediators and RANKL expression via modulating NFкB and MAPKs signaling pathways in fibroblast-like synoviocytes from adjuvant-induced arthritic rats. Immunol Res. 64:1071–1086. DOI: 10.1007/s12026-016-8794-x. PMID: 27067226.36. Feng K, Chen Z, Pengcheng L, Zhang S, Wang X. 2019; Quercetin attenuates oxidative stress-induced apoptosis via SIRT1/AMPK-mediated inhibition of ER stress in rat chondrocytes and prevents the progression of osteoarthritis in a rat model. J Cell Physiol. 234:18192–18205. DOI: 10.1002/jcp.28452. PMID: 30854676.37. Luo JS, Kammerer R, von Kleist S. 2000; Comparison of the effects of immunosuppressive factors from newly established colon carcinoma cell cultures on human lymphocyte proliferation and cytokine secretion. Tumour Biol. 21:11–20. DOI: 10.1159/000030106. PMID: 10601837.38. Park HG, Heo W, Kim SB, Kim HS, Bae GS, Chung SH, Seo HC, Kim YJ. 2011; Production of conjugated linoleic acid (CLA) by Bifidobacterium breve LMC520 and its compatibility with CLA-producing rumen bacteria. J Agric Food Chem. 59:984–988. DOI: 10.1021/jf103420q. PMID: 21192703.39. Zhang HL, Zheng L, Bian YZ, Zhang JF, Wang JH. 2011; Effects of 8-methoxypsoralen on proliferation and differentiation of cultured osteoblasts in vitro. Nat Prod Res Dev. 5:927–930.40. Ueland T, Yndestad A, Øie E, Florholmen G, Halvorsen B, Frøland SS, Simonsen S, Christensen G, Gullestad L, Aukrust P. 2005; Dysregulated osteoprotegerin/RANK ligand/RANK axis in clinical and experimental heart failure. Circulation. 111:2461–2468. DOI: 10.1161/01.CIR.0000165119.62099.14. PMID: 15883214.41. Liu G, Bi Y, Wang R, Shen B, Zhang Y, Yang H, Wang X, Liu H, Lu Y, Han F. 2013; Kinase AKT1 negatively controls neutrophil recruitment and function in mice. J Immunol. 191:2680–2690. DOI: 10.4049/jimmunol.1300736. PMID: 23904165.42. Fukai A, Kawamura N, Saito T, Oshima Y, Ikeda T, Kugimiya F, Higashikawa A, Yano F, Ogata N, Nakamura K, Chung UI, Kawaguchi H. 2010; Akt1 in murine chondrocytes controls cartilage calcification during endochondral ossification under physiologic and pathologic conditions. Arthritis Rheum. 62:826–836. DOI: 10.1002/art.27296. PMID: 20187155.43. Saklatvala J. 1986; Tumour necrosis factor alpha stimulates resorption and inhibits synthesis of proteoglycan in cartilage. Nature. 322:547–549. DOI: 10.1038/322547a0. PMID: 3736671. PMCID: PMC7095107.44. Séguin CA, Bernier SM. 2003; TNFalpha suppresses link protein and type II collagen expression in chondrocytes: role of MEK1/2 and NF-kappaB signaling pathways. J Cell Physiol. 197:356–369. DOI: 10.1002/jcp.10371. PMID: 14566965.45. Haag J, Gebhard PM, Aigner T. 2008; SOX gene expression in human osteoarthritic cartilage. Pathobiology. 75:195–199. DOI: 10.1159/000124980. PMID: 18550917.46. Li Z, Chen S, Chen S, Huang D, Ma K, Shao Z. 2019; Moderate activation of Wnt/β-catenin signaling promotes the survival of rat nucleus pulposus cells via regulating apoptosis, autophagy, and senescence. J Cell Biochem. 120:12519–12533. DOI: 10.1002/jcb.28518. PMID: 31016779.47. Wang J, Wang Y, Zhang H, Gao W, Lu M, Liu W, Li Y, Yin Z. 2020; Forkhead box C1 promotes the pathology of osteoarthritis by upregulating β-catenin in synovial fibroblasts. FEBS J. 287:3065–3087. DOI: 10.1111/febs.15178. PMID: 31837247.48. Chen B, Deng Y, Tan Y, Qin J, Chen LB. 2014; Association between severity of knee osteoarthritis and serum and synovial fluid interleukin 17 concentrations. J Int Med Res. 42:138–144. DOI: 10.1177/0300060513501751. PMID: 24319050.49. Chen Y, Shou K, Gong C, Yang H, Yang Y, Bao T. 2018; Anti-inflammatory effect of geniposide on osteoarthritis by suppressing the activation of p38 MAPK signaling pathway. Biomed Res Int. 2018:8384576. Erratum in: Biomed Res Int. 2022;2022:9814323. DOI: 10.1155/2018/8384576. PMID: 29682561. PMCID: PMC5846349.50. Shi J, Zhang C, Yi Z, Lan C. 2016; Explore the variation of MMP3, JNK, p38 MAPKs, and autophagy at the early stage of osteoarthritis. IUBMB Life. 68:293–302. DOI: 10.1002/iub.1482. PMID: 26873249.51. Morris R, Kershaw NJ, Babon JJ. 2018; The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci. 27:1984–2009. DOI: 10.1002/pro.3519. PMID: 30267440. PMCID: PMC6237706.52. Leisengang S, Gluding D, Hörster J, Peek V, Ott D, Rummel C, Schmidt MJ. 2022; Expression of adipokines and adipocytokines by epidural adipose tissue in cauda equina syndrome in dogs. J Vet Intern Med. 36:1373–1381. DOI: 10.1111/jvim.16483. PMID: 35838307. PMCID: PMC9308421. PMID: 1f63b64ed3834813b4b8c1b80a1d11eb.53. Chen H, Wu G, Sun Q, Dong Y, Zhao H. 2016; Hyperbaric oxygen protects mandibular condylar chondrocytes from interleukin-1β-induced apoptosis via the PI3K/AKT signaling pathway. Am J Transl Res. 8:5108–5117. PMID: 27904712. PMCID: PMC5126354.54. Zhang Q, Lai S, Hou X, Cao W, Zhang Y, Zhang Z. 2018; Protective effects of PI3K/Akt signal pathway induced cell autophagy in rat knee joint cartilage injury. Am J Transl Res. 10:762–770. PMID: 29636866. PMCID: PMC5883117.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Network pharmacology prediction to discover the potential pharmacological action mechanism of Rhizoma Dioscoreae for liver regeneration
- Application of Musculoskeletal Ultrasound in Osteoarthritis
- Experimental Animal Models of Portal Hypertension
- Preliminary Study of Experimental Neural Network Model in the Nervous System
- Behind the Naming of Herbal Section as the Decoction Section in Treasured Mirror of Eastern Medicine