Diabetes Metab J.
2023 Sep;47(5):668-681. 10.4093/dmj.2022.0342.
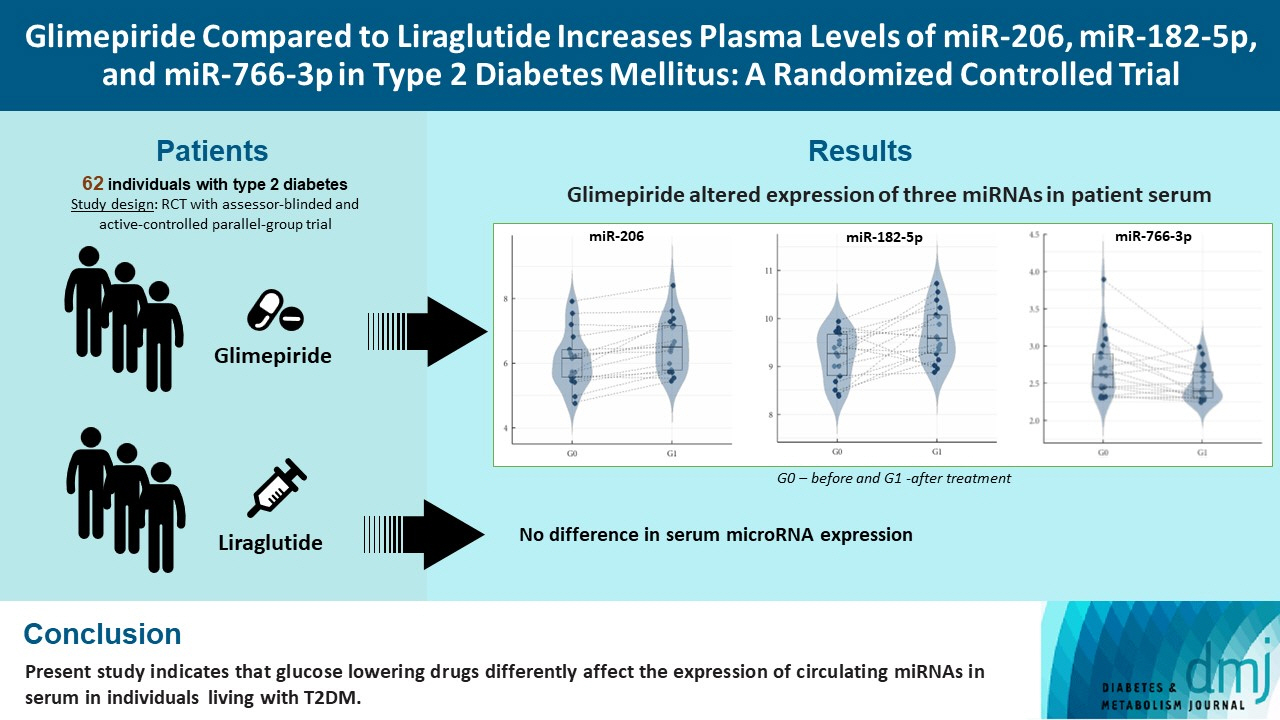
Glimepiride Compared to Liraglutide Increases Plasma Levels of miR-206, miR-182-5p, and miR-766-3p in Type 2 Diabetes Mellitus: A Randomized Controlled Trial
- Affiliations
-
- 1Life Science Center, Örebro University, School of Science and Technology, Örebro, Sweden
- 2Department of Clinical Research Laboratory, 3Inflammatory Response and Infection Susceptibility Center (iRiSC), Faculty of Medicine and Health, Örebro University, Örebro, Sweden
- 3Karolinska Institutet, Department of Clinical Science and Education, Södersjukhuset, Stockholm, Sweden
- 4School of Medical Sciences, Faculty of Medicine and Health, Örebro University, Örebro, Sweden
- KMID: 2546125
- DOI: http://doi.org/10.4093/dmj.2022.0342
Abstract
- Background
Diabetes is a chronic disease with several long-term complications. Several glucose-lowering drugs are used to treat type 2 diabetes mellitus (T2DM), e.g., glimepiride and liraglutide, in which both having different modes of action. Circulating microRNAs (miRNAs) are suggested as potential biomarkers that are associated with the disease development and the effects of the treatment. In the current study we evaluated the effect of glimepiride, liraglutide on the expression of the circulating miRNAs.
Methods
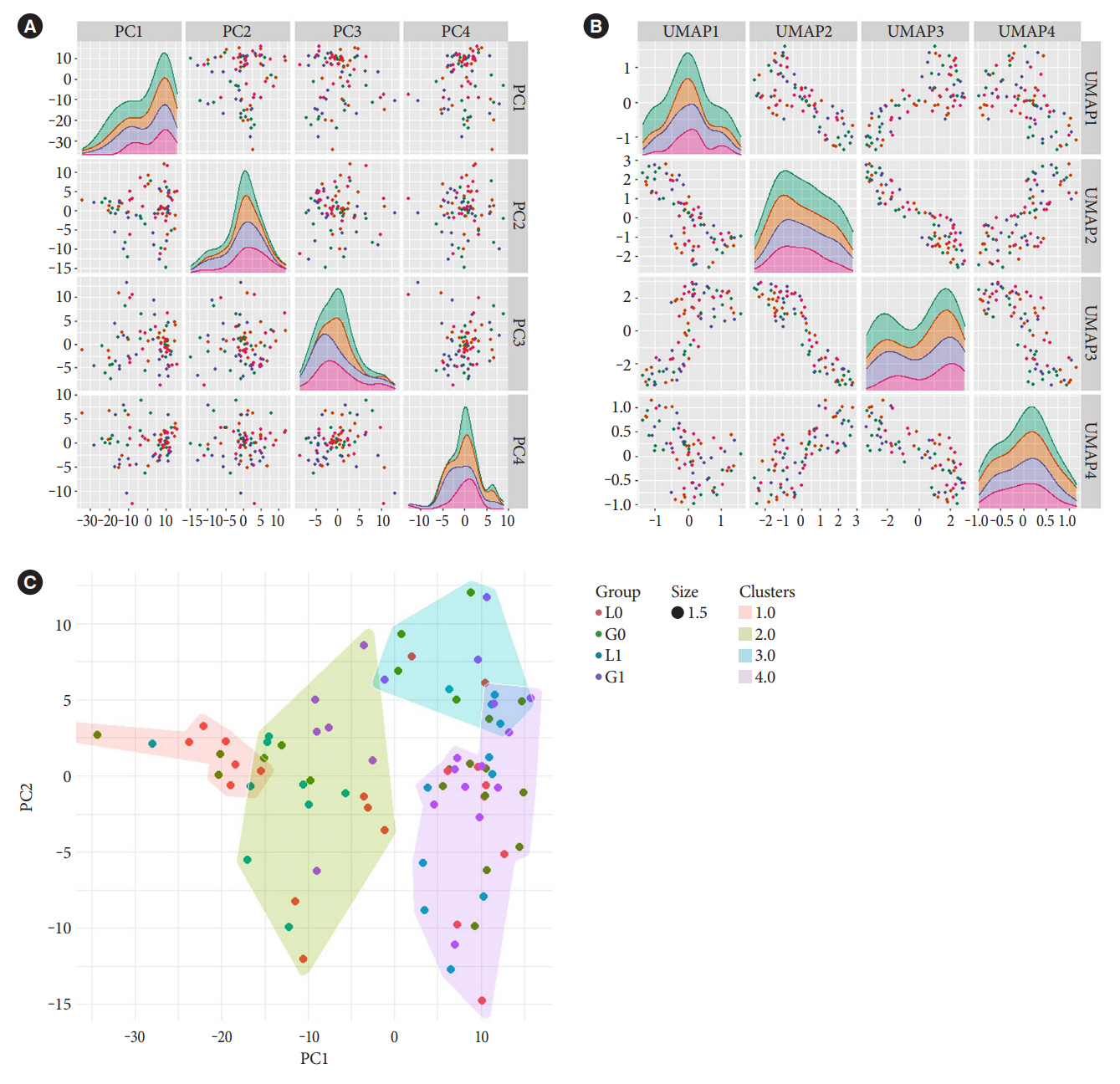
The present study is a post hoc trial from a previously randomized control trial comparing liraglutide versus glimepiride both in combination with metformin in subjects with T2DM, and subclinical heart failure. miRNAs were determined in the subjects’ serum samples with next generation sequencing. Expression patterns of the circulating miRNAs were analyzed using bioinformatic univariate and multivariate analyses (clinical trial registration: NCT01425580).
Results
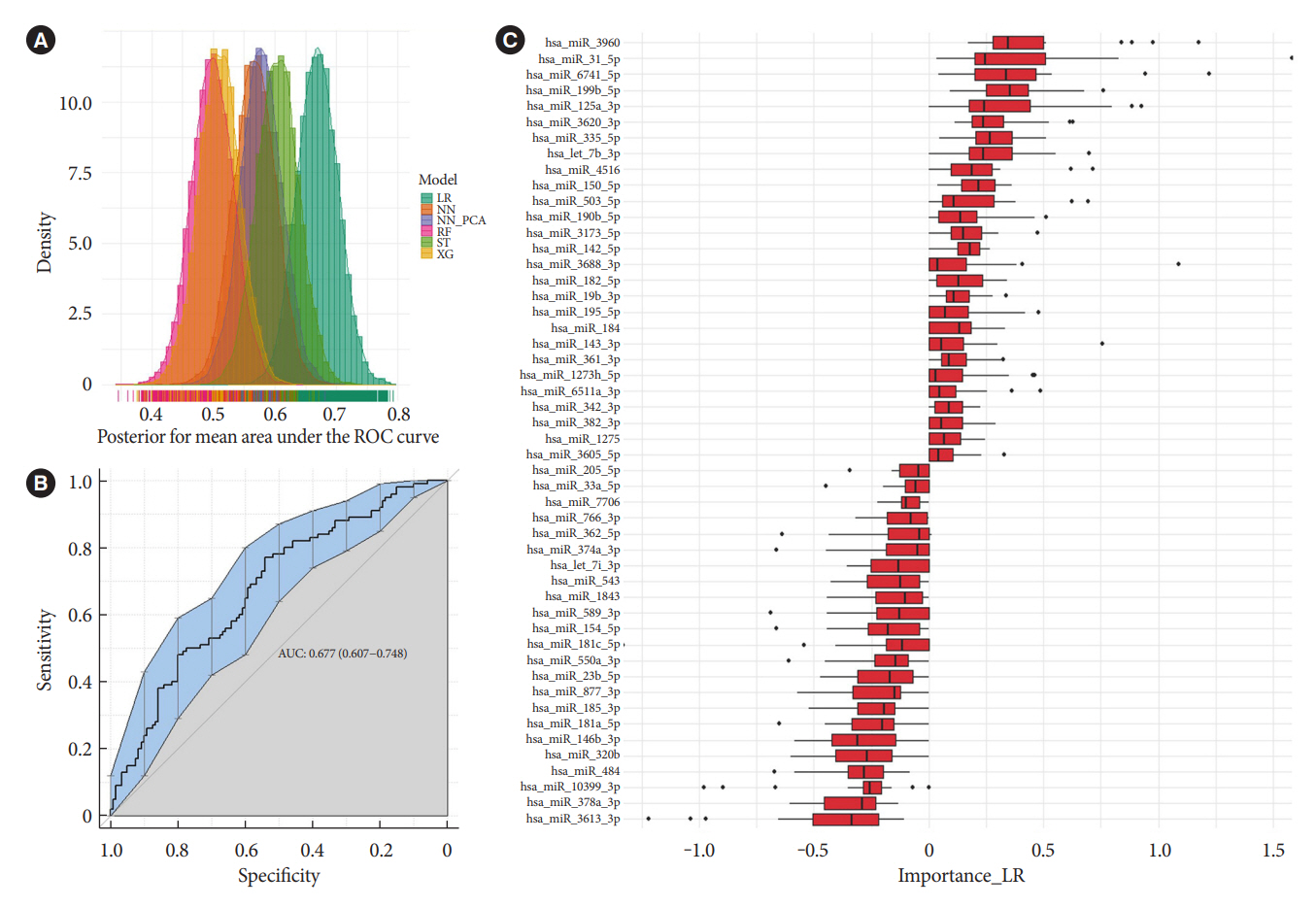
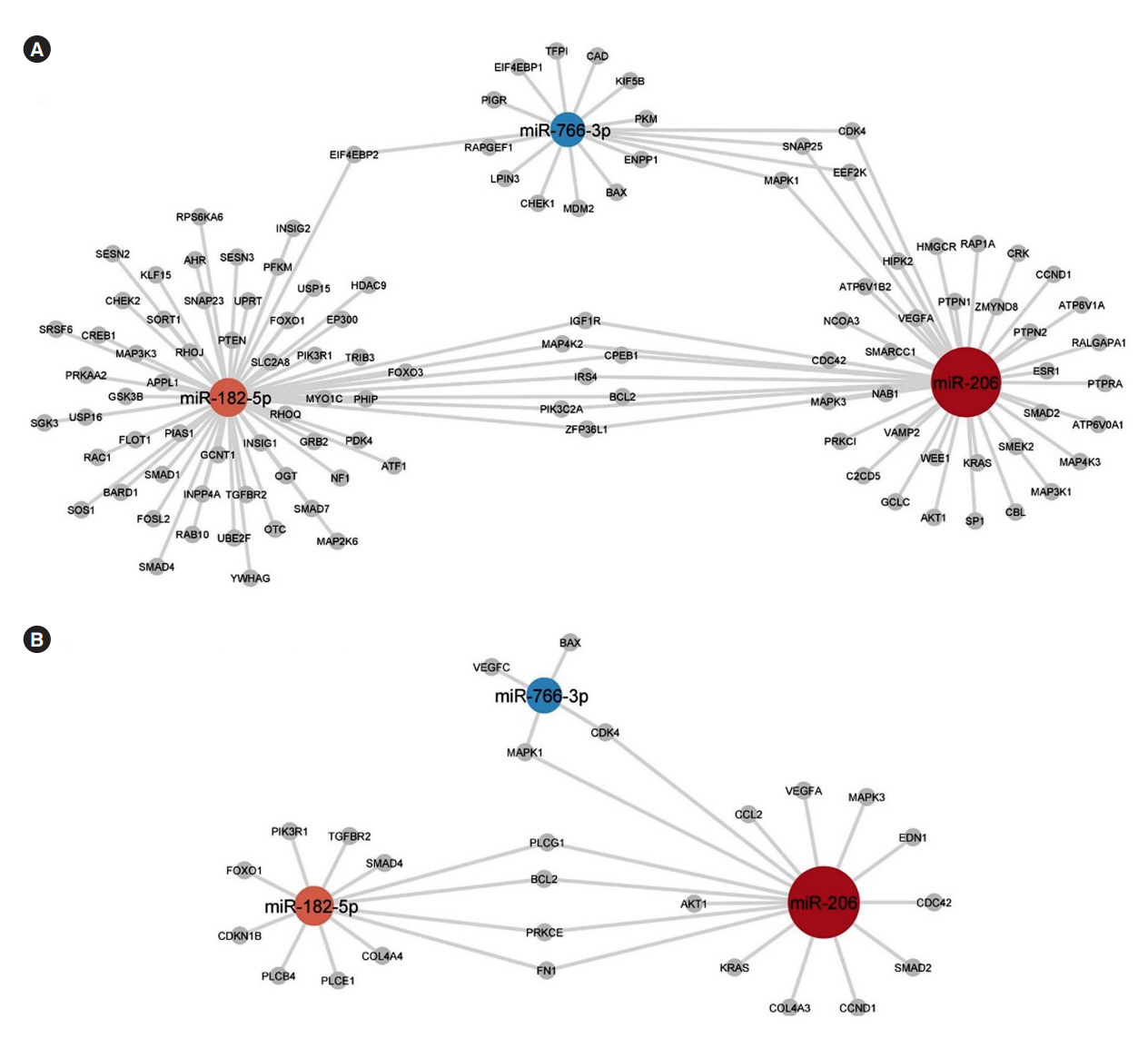
Univariate analyses show that treatment with glimepiride altered expression of three miRNAs in patient serum, miR-206, miR-182-5p, and miR-766-3p. Both miR-182-5p and miR-766-3p were also picked up among the top contributing miRNAs with penalized regularised logistic regressions (Lasso). The highest-ranked miRNAs with respect to Lasso coefficients were miR-3960, miR-31-5p, miR-3613-3p, and miR-378a-3p. Liraglutide treatment did not significantly influence levels of circulating miRNAs.
Conclusion
Present study indicates that glucose-lowering drugs differently affect the expression of circulating miRNAs in serum in individuals with T2DM. More studies are required to investigate possible mechanisms by which glimepiride is affecting the expression of circulating miRNAs.
Figure
Cited by 2 articles
-
Glimepiride Compared to Liraglutide Increases Plasma Levels of miR-206, miR-182-5p, and miR-766-3p in Type 2 Diabetes Mellitus: A Randomized Controlled Trial (
Diabetes Metab J 2023;47:668-81)
Nikolai N. Scherbak, Robert Kruse, Thomas Nyström, Johan Jendle
Diabetes Metab J. 2023;47(6):882-883. doi: 10.4093/dmj.2023.0355.Glimepiride Compared to Liraglutide Increases Plasma Levels of miR-206, miR-182-5p, and miR-766-3p in Type 2 Diabetes Mellitus: A Randomized Controlled Trial (
Diabetes Metab J 2023;47:668-81)
Da Young Lee
Diabetes Metab J. 2023;47(6):879-881. doi: 10.4093/dmj.2023.0328.
Reference
-
1. NCD Risk Factor Collaboration (NCD-RisC). Worldwide trends in diabetes since 1980: a pooled analysis of 751 population-based studies with 4.4 million participants. Lancet. 2016; 387:1513–30.2. Holman RR, Paul SK, Bethel MA, Matthews DR, Neil HA. 10-Year follow-up of intensive glucose control in type 2 diabetes. N Engl J Med. 2008; 359:1577–89.3. Rao Kondapally Seshasai S, Kaptoge S, Thompson A, Di Angelantonio E, Gao P, Sarwar N, et al. Diabetes mellitus, fasting glucose, and risk of cause-specific death. N Engl J Med. 2011; 364:829–41.4. UK Prospective Diabetes Study Group. Tight blood pressure control and risk of macrovascular and microvascular complications in type 2 diabetes: UKPDS 38. BMJ. 1998; 317:703–13.5. Colhoun HM, Betteridge DJ, Durrington PN, Hitman GA, Neil HA, Livingstone SJ, et al. Primary prevention of cardiovascular disease with atorvastatin in type 2 diabetes in the Collaborative Atorvastatin Diabetes Study (CARDS): multicentre randomised placebo-controlled trial. Lancet. 2004; 364:685–96.6. Grant RW, Pirraglia PA, Meigs JB, Singer DE. Trends in complexity of diabetes care in the United States from 1991 to 2000. Arch Intern Med. 2004; 164:1134–9.7. American Diabetes Association. Standards of medical care in diabetes: 2014. Diabetes Care. 2014; 37 Suppl 1:S14–80.8. Swedish National Diabetes Register (NDR) Centre of Registers Västra Götaland. Nation-wide results 1996-2020. Gothenburg: NDR;2020. Available from: https://www.ndr.nu/pdfs/NationWideResults_1996-2020.pdf.9. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008; 105:10513–8.10. Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010; 107:810–7.11. De Smaele E, Ferretti E, Gulino A. MicroRNAs as biomarkers for CNS cancer and other disorders. Brain Res. 2010; 1338:100–11.12. Ciccacci C, Morganti R, Di Fusco D, D’Amato C, Cacciotti L, Greco C, et al. Common polymorphisms in MIR146a, MIR128a and MIR27a genes contribute to neuropathy susceptibility in type 2 diabetes. Acta Diabetol. 2014; 51:663–71.13. Barbagallo D, Piro S, Condorelli AG, Mascali LG, Urbano F, Parrinello N, et al. miR-296-3p, miR-298-5p and their downstream networks are causally involved in the higher resistance of mammalian pancreatic α cells to cytokine-induced apoptosis as compared to β cells. BMC Genomics. 2013; 14:62.14. Romaine SP, Tomaszewski M, Condorelli G, Samani NJ. MicroRNAs in cardiovascular disease: an introduction for clinicians. Heart. 2015; 101:921–8.15. Guay C, Regazzi R. New emerging tasks for microRNAs in the control of β-cell activities. Biochim Biophys Acta. 2016; 1861(12 Pt B):2121–9.16. Sebastiani G, Po A, Miele E, Ventriglia G, Ceccarelli E, Bugliani M, et al. MicroRNA-124a is hyperexpressed in type 2 diabetic human pancreatic islets and negatively regulates insulin secretion. Acta Diabetol. 2015; 52:523–30.17. Jendle J, Fang X, Cao Y, Bojo L, Nilsson BK, Hedberg F, et al. Effects on repetitive 24-hour ambulatory blood pressure in subjects with type II diabetes randomized to liraglutide or glimepiride treatment both in combination with metformin: a randomized open parallel-group study. J Am Soc Hypertens. 2018; 12:346–55.18. Nystrom T, Santos-Pardo I, Fang X, Cao Y, Hedberg F, Jendle J. Heart rate variability in type 2 diabetic subjects randomized to liraglutide or glimepiride treatment, both in combination with metformin: a randomized, open, parallel-group study. Endocrinol Diabetes Metab. 2019; 2:e00058.19. Nystrom T, Santos-Pardo I, Hedberg F, Wardell J, Witt N, Cao Y, et al. Effects on subclinical heart failure in type 2 diabetic subjects on liraglutide treatment vs. glimepiride both in combination with metformin: a randomized open parallel-group study. Front Endocrinol (Lausanne). 2017; 8:325.20. McElreath R. Statistical rethinking: a Bayesian course with examples in R and Stan. 2nd ed. London: Chapman and Hall/CRC;2020. p. 594.21. Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004; 5:R13.22. Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci U S A. 2006; 103:8721–6.23. Yan Y, Dang H, Zhang X, Wang X, Liu X. The protective role of MiR-206 in regulating cardiomyocytes apoptosis induced by ischemic injury by targeting PTP1B. Biosci Rep. 2020; 40:BSR20191000.24. Shan ZX, Lin QX, Deng CY, Zhu JN, Mai LP, Liu JL, et al. miR-1/miR-206 regulate Hsp60 expression contributing to glucose-mediated apoptosis in cardiomyocytes. FEBS Lett. 2010; 584:3592–600.25. Vinod M, Patankar JV, Sachdev V, Frank S, Graier WF, Kratky D, et al. MiR-206 is expressed in pancreatic islets and regulates glucokinase activity. Am J Physiol Endocrinol Metab. 2016; 311:E175–85.26. Joglekar MV, Wong WK, Maynard CL, Umrani MR, Martin D, Loudovaris T, et al. Expression of miR-206 in human islets and its role in glucokinase regulation. Am J Physiol Endocrinol Metab. 2018; 315:E634–7.27. Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008; 18:997–1006.28. Raffort J, Hinault C, Dumortier O, Van Obberghen E. Circulating microRNAs and diabetes: potential applications in medical practice. Diabetologia. 2015; 58:1978–92.29. Peters LJ, Biessen EA, Hohl M, Weber C, van der Vorst EP, Santovito D. Small things matter: relevance of microRNAs in cardiovascular disease. Front Physiol. 2020; 11:793.30. Gomes CP, Oliveira GP, Madrid B, Almeida JA, Franco OL, Pereira RW. Circulating miR-1, miR-133a, and miR-206 levels are increased after a half-marathon run. Biomarkers. 2014; 19:585–9.31. Li X, Li Y, Zhao L, Zhang D, Yao X, Zhang H, et al. Circulating muscle-specific miRNAs in Duchenne muscular dystrophy patients. Mol Ther Nucleic Acids. 2014; 3:e177.32. Toivonen JM, Manzano R, Olivan S, Zaragoza P, Garcia-Redondo A, Osta R. MicroRNA-206: a potential circulating biomarker candidate for amyotrophic lateral sclerosis. PLoS One. 2014; 9:e89065.33. Heinemann FG, Tolkach Y, Deng M, Schmidt D, Perner S, Kristiansen G, et al. Serum miR-122-5p and miR-206 expression: non-invasive prognostic biomarkers for renal cell carcinoma. Clin Epigenetics. 2018; 10:11.34. Liu X, Zheng W, Zhang X, Dong M, Sun G. The diagnostic and prognostic value of serum miR-206 in colorectal cancer. Int J Clin Exp Pathol. 2017; 10:7528–33.35. Jin P, Gu W, Lai Y, Zheng W, Zhou Q, Wu X. The circulating microRNA-206 level predicts the severity of pulmonary hypertension in patients with left heart diseases. Cell Physiol Biochem. 2017; 41:2150–60.36. Infante T, Forte E, Punzo B, Cademartiri F, Cavaliere C, Soricelli A, et al. Correlation of circulating miR-765, miR-93-5p, and miR-433-3p to obstructive coronary heart disease evaluated by cardiac computed tomography. Am J Cardiol. 2019; 124:176–82.37. Kim M, Zhang X. The profiling and role of miRNAs in diabetes mellitus. J Diabetes Clin Res. 2019; 1:5–23.38. Wu H, Zhang T, Pan F, Steer CJ, Li Z, Chen X, et al. MicroRNA-206 prevents hepatosteatosis and hyperglycemia by facilitating insulin signaling and impairing lipogenesis. J Hepatol. 2017; 66:816–24.39. Zhu L, Chen T, Ye W, Wang JY, Zhou JP, Li ZY, et al. Circulating miR-182-5p and miR-5187-5p as biomarkers for the diagnosis of unprotected left main coronary artery disease. J Thorac Dis. 2019; 11:1799–808.40. Weale CJ, Matshazi DM, Davids SF, Raghubeer S, Erasmus RT, Kengne AP, et al. Circulating miR-30a-5p and miR-182-5p in prediabetes and screen-detected diabetes mellitus. Diabetes Metab Syndr Obes. 2020; 13:5037–47.41. Weale CJ, Matshazi DM, Davids SF, Raghubeer S, Erasmus RT, Kengne AP, et al. Expression profiles of circulating microRNAs in South African type 2 diabetic individuals on treatment. Front Genet. 2021; 12:702410.42. Karolina DS, Armugam A, Tavintharan S, Wong MT, Lim SC, Sum CF, et al. MicroRNA 144 impairs insulin signaling by inhibiting the expression of insulin receptor substrate 1 in type 2 diabetes mellitus. PLoS One. 2011; 6:e22839.43. Liu S, Lin Z, Zheng Z, Rao W, Lin Y, Chen H, et al. Serum exosomal microRNA-766-3p expression is associated with poor prognosis of esophageal squamous cell carcinoma. Cancer Sci. 2020; 111:3881–92.44. Owczarz M, Polosak J, Domaszewska-Szostek A, Kolodziej P, Kurylowicz A, Puzianowska-Kuznicka M. Age-related epigenetic drift deregulates SIRT6 expression and affects its downstream genes in human peripheral blood mononuclear cells. Epigenetics. 2020; 15:1336–47.45. Hayakawa K, Kawasaki M, Hirai T, Yoshida Y, Tsushima H, Fujishiro M, et al. MicroRNA-766-3p contributes to anti-inflammatory responses through the indirect inhibition of NF-κB signaling. Int J Mol Sci. 2019; 20:809.46. Yang ZM, Chen LH, Hong M, Chen YY, Yang XR, Tang SM, et al. Serum microRNA profiling and bioinformatics analysis of patients with type 2 diabetes mellitus in a Chinese population. Mol Med Rep. 2017; 15:2143–53.47. Olioso D, Dauriz M, Bacchi E, Negri C, Santi L, Bonora E, et al. Effects of aerobic and resistance training on circulating microRNA expression profile in subjects with type 2 diabetes. J Clin Endocrinol Metab. 2019; 104:1119–30.48. Cao Z, Yao F, Lang Y, Feng X. Elevated circulating LINC-P21 serves as a diagnostic biomarker of type 2 diabetes mellitus and regulates pancreatic β-cell function by sponging miR-766-3p to upregulate NR3C2. Exp Clin Endocrinol Diabetes. 2022; 130:156–64.49. Demirsoy lH, Ertural DY, Balci S, Cınkır U, Sezer K, Tamer L, et al. Profiles of circulating MiRNAs following metformin treatment in patients with type 2 diabetes. J Med Biochem. 2018; 37:499–506.50. Ortega FJ, Mercader JM, Moreno-Navarrete JM, Rovira O, Guerra E, Esteve E, et al. Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes Care. 2014; 37:1375–83.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Glimepiride Compared to Liraglutide Increases Plasma Levels of miR-206, miR-182-5p, and miR-766-3p in Type 2 Diabetes Mellitus: A Randomized Controlled Trial (Diabetes Metab J 2023;47:668-81)
- Glimepiride Compared to Liraglutide Increases Plasma Levels of miR-206, miR-182-5p, and miR-766-3p in Type 2 Diabetes Mellitus: A Randomized Controlled Trial (Diabetes Metab J 2023;47:668-81)
- Combined Detection of Serum MiR-221-3p and MiR-122-5p Expression in Diagnosis and Prognosis of Gastric Cancer
- miR-132 and miR-942 Expression Levels in Children with Attention Deficit and Hyperactivity Disorder: A Controlled Study
- Expression of specific microRNAs in tissue and plasma in colorectal cancer