Clin Exp Otorhinolaryngol.
2023 Feb;16(1):75-86. 10.21053/ceo.2022.01480.
SERPINE1 as an Independent Prognostic Marker and Therapeutic Target for Nicotine-Related Oral Carcinoma
- Affiliations
-
- 1Department of Otorhinolaryngology-Head and Neck Surgery, Affiliated Hospital of Guizhou Medical University, Guiyang, China
- KMID: 2539771
- DOI: http://doi.org/10.21053/ceo.2022.01480
Abstract
Objectives
. Nicotine is an ingredient of tobacco, and exposure to nicotine increases the risks of various cancers, including oral cancer. Previous studies have focused on the addictive properties of nicotine, but its carcinogenic mechanism has rarely been studied. We aimed to explore the key genes in the process through which nicotine promotes the occurrence and development of oral cancer via data mining and experimental verification.
Methods
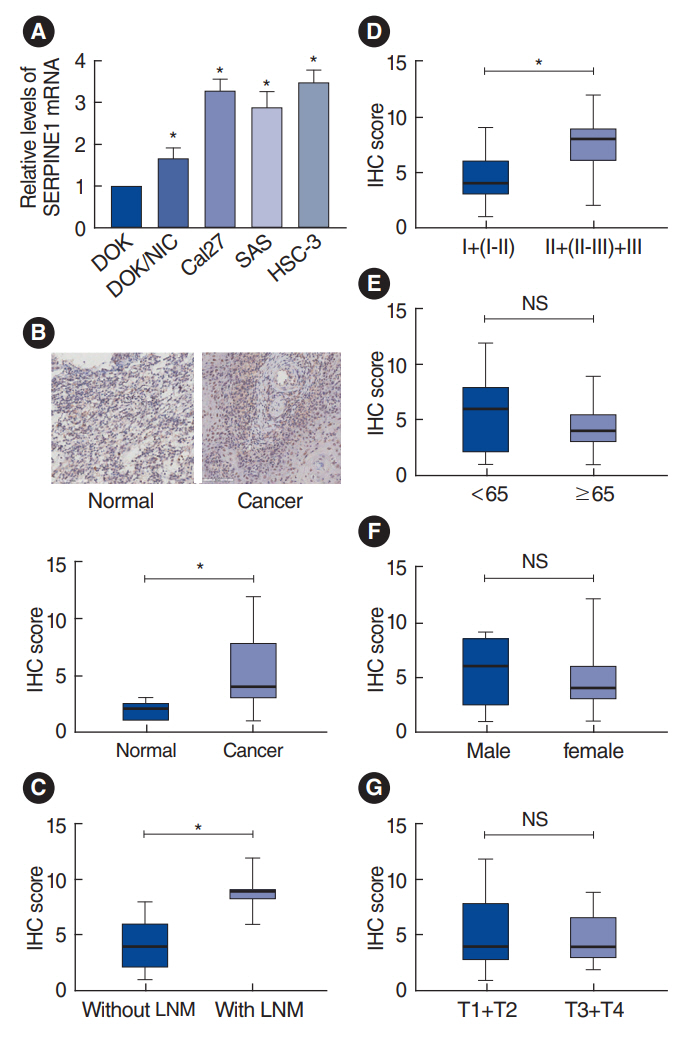
. This study involved three parts. First, key genes related to nicotine-related oral cancer were screened through data mining; second, the expression and clinical significance of a key gene in oral cancer tissues were verified by bioinformatics. Finally, the expression and clinical significance of the key gene in oral cancer were histologically investigated, and the effects of its expression on cell proliferation, invasion, and drug resistance were cytologically assessed.
Results
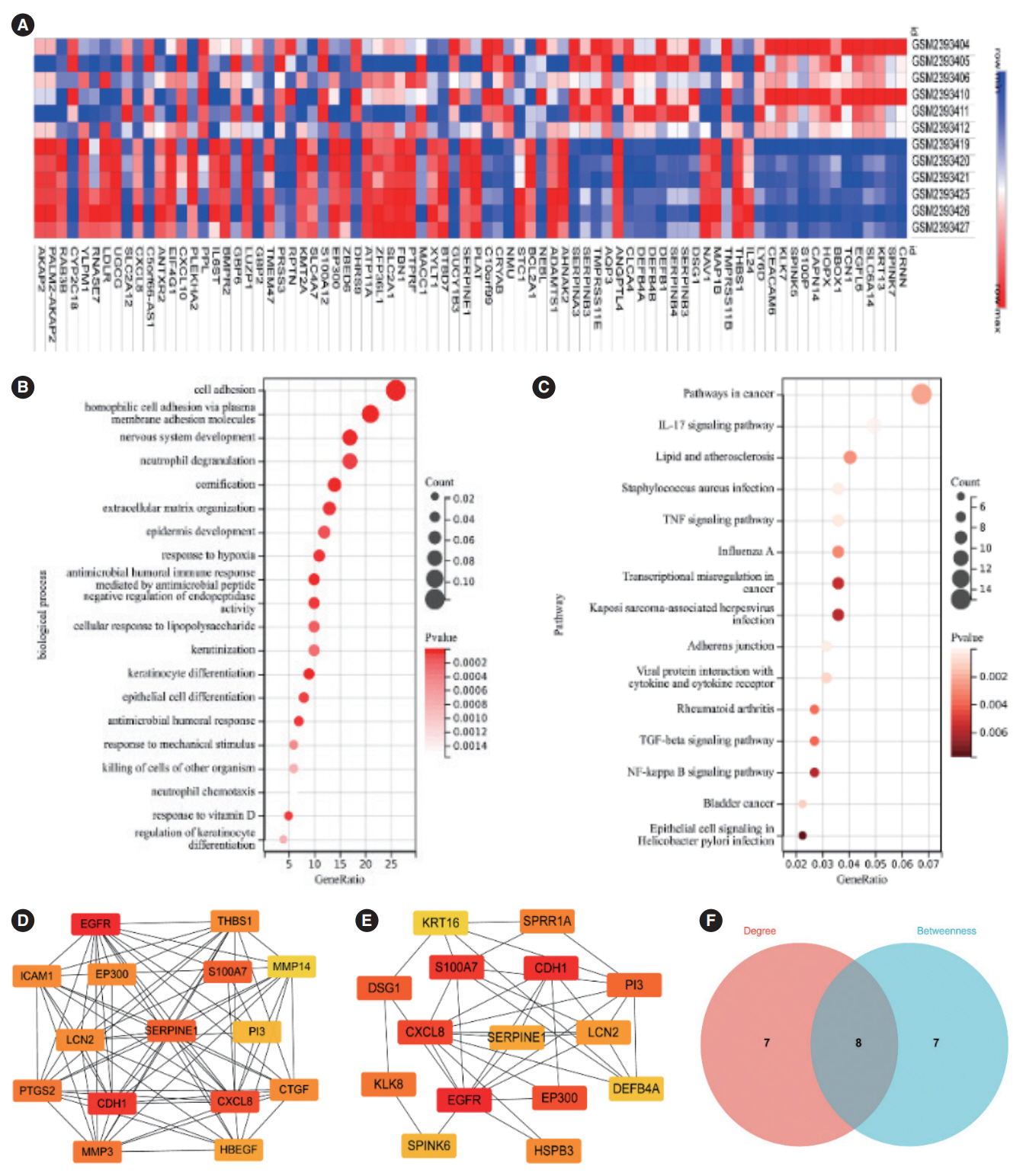
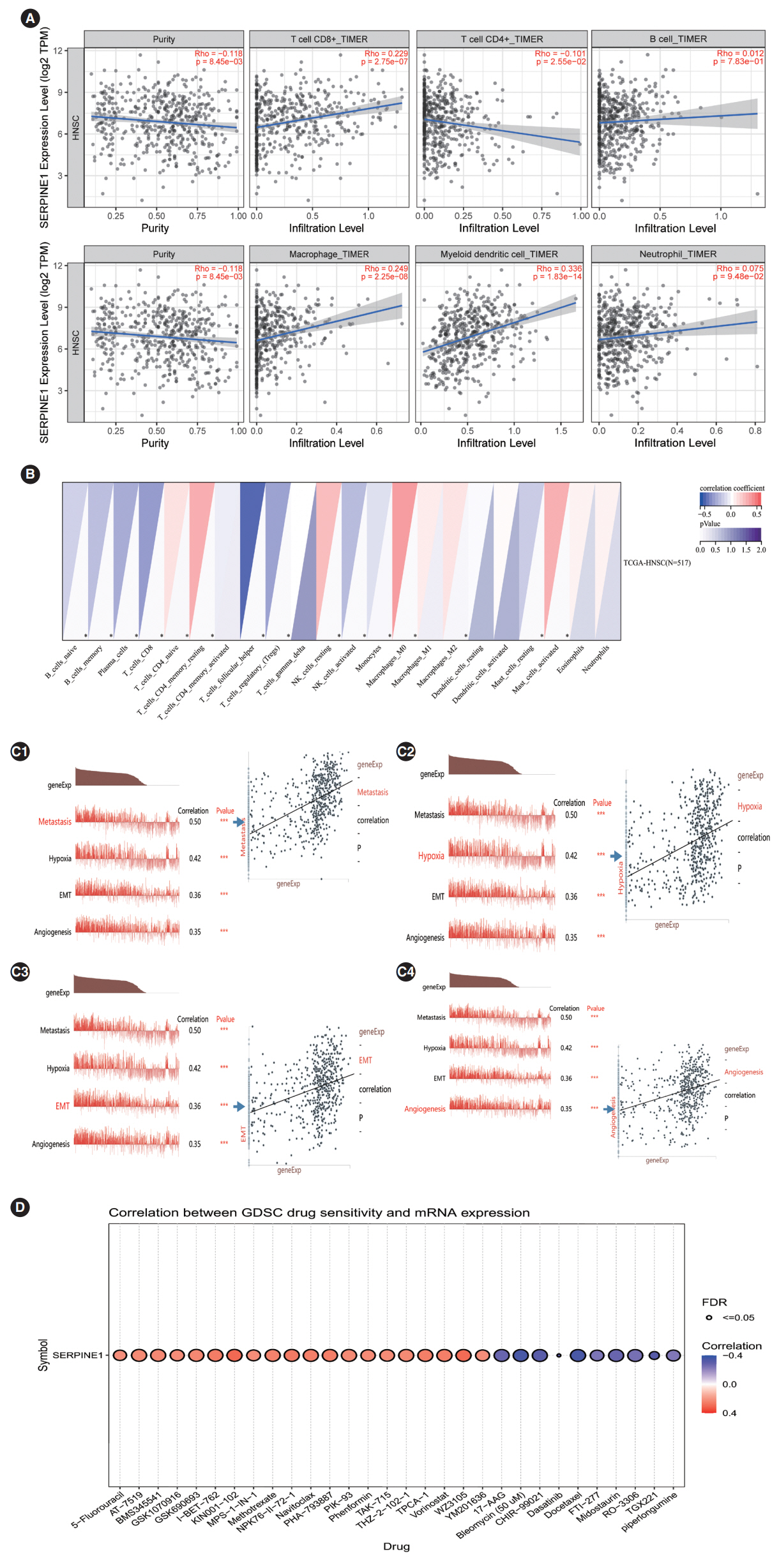
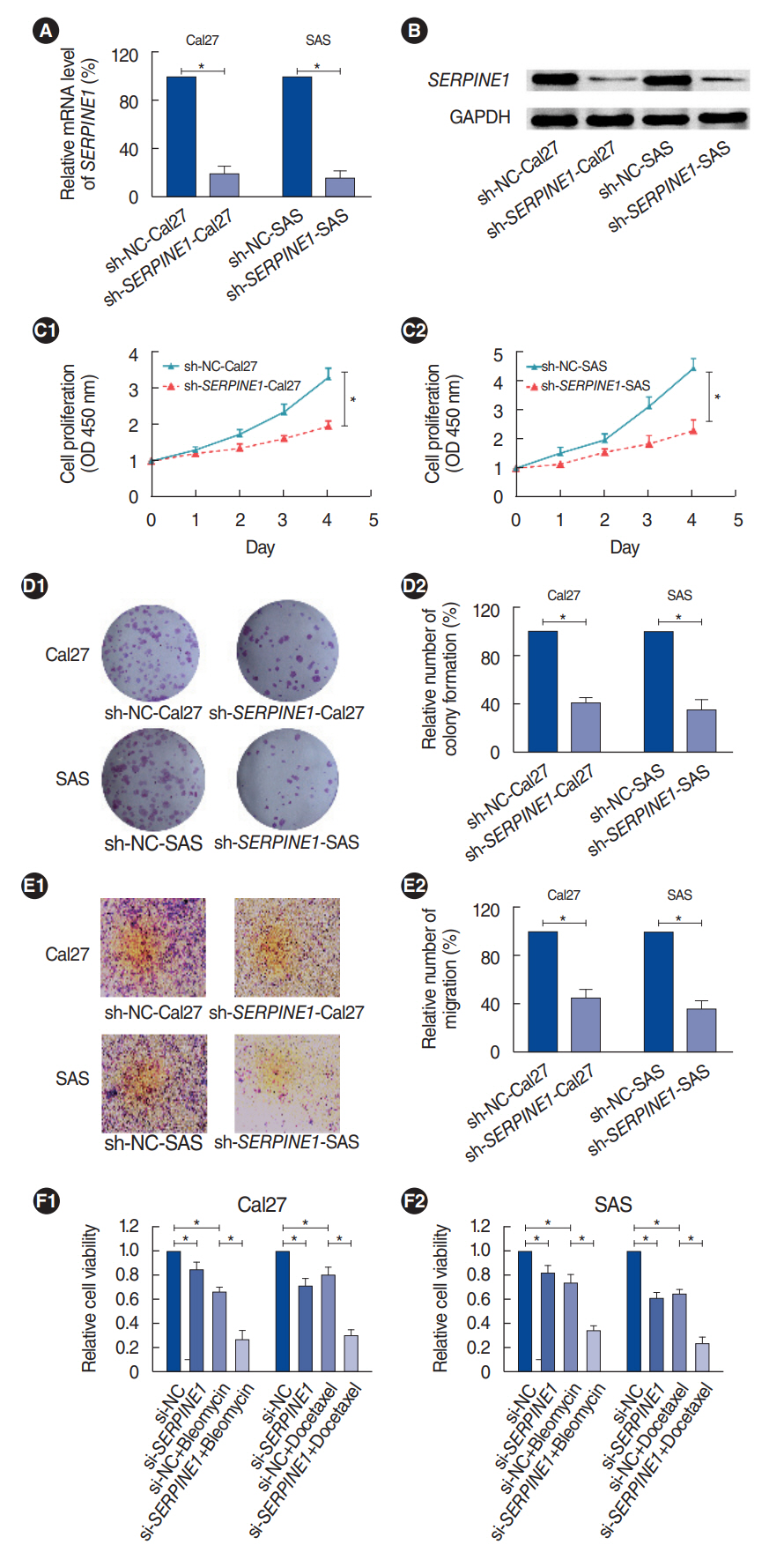
. SERPINE1 was identified as the key gene, which was upregulated in nicotine-treated oral cells and may be an independent prognostic factor for oral cancer. SERPINE1 was enriched in various pathways, such as the tumor necrosis factor and apelin pathways, and was related to the infiltration of macrophages, CD4+T cells, and CD8+T cells. Overexpression of SERPINE1 was associated with N staging and may be involved in hypoxia, angiogenesis, and metastasis. Knockdown of SERPINE1 in oral cancer cells resulted in weakened cell proliferation and invasion ability and increased sensitivity to bleomycin and docetaxel.
Conclusion
. This study revealed SERPINE1 as a key gene for nicotine-related oral cancer, indicating that SERPINE1 may be a novel prognostic indicator and therapeutic target for oral carcinoma.
Figure
Reference
-
1. Rivera C. Essentials of oral cancer. Int J Clin Exp Pathol. 2015; Sep. 8(9):11884–94.2. Auperin A. Epidemiology of head and neck cancers: an update. Curr Opin Oncol. 2020; May. 32(3):178–86.
Article3. Onor IO, Stirling DL, Williams SR, Bediako D, Borghol A, Harris MB, et al. Clinical effects of cigarette smoking: epidemiologic impact and review of pharmacotherapy options. Int J Environ Res Public Health. 2017; Sep. 14(10):1147.
Article4. Alkhatib R, Alkhatib B, Abdo N. Impact of exogenous nicotine on the morphological, physio-biochemical, and anatomical characteristics in Capsicum annuum. Int J Phytoremediation. 2022; 24(6):666–74.5. Murphy SE. Biochemistry of nicotine metabolism and its relevance to lung cancer. J Biol Chem. 2021; Jan-Jun. 296:100722.
Article6. Dang N, Meng X, Song H. Nicotinic acetylcholine receptors and cancer. Biomed Rep. 2016; May. 4(5):515–8.
Article7. Chen J, Cheuk IW, Shin VY, Kwong A. Acetylcholine receptors: Key players in cancer development. Surg Oncol. 2019; Dec. 31:46–53.
Article8. Shimizu R, Ibaragi S, Eguchi T, Kuwajima D, Kodama S, Nishioka T, et al. Nicotine promotes lymph node metastasis and cetuximab resistance in head and neck squamous cell carcinoma. Int J Oncol. 2019; Jan. 54(1):283–94.
Article9. Zhang Y, Sun Y, Jia Y, Zhang Q, Zhu P, Ma X. α5-nAChR and survivin: two potential biological targets in lung adenocarcinoma. J Cell Physiol. 2021; Mar. 236(3):1787–97.
Article10. Qi M, Li L, Tang X, Lu Y, Wang M, Yang J, et al. Nicotine promotes the development of oral leukoplakia via regulating peroxiredoxin 1 and its binding proteins. Braz J Med Biol Res. 2021; May. 54(9):e10931.
Article11. Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets: update. Nucleic Acids Res. 2013; Jan. 41(Database issue):D991–5.12. Yuan H, Yan M, Zhang G, Liu W, Deng C, Liao G, et al. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 2019; Jan. 47(D1):D900–8.
Article13. Li X, Yang A, Wen P, Yuan Y, Xiao Z, Shi H, et al. Nuclear receptor subfamily 3 group c member 2 (NR3C2) is downregulated due to hypermethylation and plays a tumor-suppressive role in colon cancer. Mol Cell Biochem. 2022; Nov. 477(11):2669–79.
Article14. Feng S, Yuan W, Sun Z, Guo X, Ling J, Chang A, et al. SPP1 as a key gene in the lymph node metastasis and a potential predictor of poor prognosis in head and neck carcinoma. J Oral Pathol Med. 2022; Aug. 51(7):620–9.15. Chang YW, Singh KP. Nicotine-induced oxidative stress contributes to EMT and stemness during neoplastic transformation through epigenetic modifications in human kidney epithelial cells. Toxicol Appl Pharmacol. 2019; Jul. 374:65–76.
Article16. Woo S, Gao H, Henderson D, Zacharias W, Liu G, Tran QT, et al. AKR1C1 as a biomarker for differentiating the biological effects of combustible from non-combustible tobacco products. Genes (Basel). 2017; May. 8(5):132.
Article17. Zhang T, Yu H, Dai X, Zhang X. CMTM6 and CMTM4 as two novel regulators of PD-L1 modulate the tumor microenvironment. Front Immunol. 2022; Jul. 13:971428.
Article18. Chen TY, Zhou M, Lin MQ, Liang ST, Yan Y, Wang SM, et al. Research progress on the SERPINE1 protein and chronic inflammatory diseases of the upper respiratory tract: a literature review. Int Arch Allergy Immunol. 2021; 182(11):1097–102.
Article19. Hu B, Chen Z, Wang X, Chen F, Song Z, Cao C. MicroRNA-148a-3p directly targets SERPINE1 to suppress EMT-Mediated colon adenocarcinoma progression. Cancer Manag Res. 2021; Aug. 13:6349–62.
Article20. Dai Y, Wang Z, Yan E, Li J, Ge H, Xiao N, et al. Development of a novel signature derived from single cell RNA-sequencing for preoperative prediction of lymph node metastasis in head and neck squamous cell carcinoma. Head Neck. 2022; Oct. 44(10):2171–80.21. Sugiura K, Nakajima S, Kato I, Okubo-Sato M, Nakazawa Y, Mitsudo K, et al. Hypoxia and CD11b+ cell influx are strongly associated with lymph node metastasis of oral cancer. Anticancer Res. 2020; Dec. 40(12):6845–52.
Article22. Li Jp, Liu YJ, Zeng SH, Gao HJ, Chen YG, Zou X. Identification of COX4I2 as a hypoxia-associated gene acting through FGF1 to promote EMT and angiogenesis in CRC. Cell Mol Biol Lett. 2022; Sep. 27(1):76.23. Teng B, Xie C, Zhao Y, Zeng Q, Zhan F, Feng Y, et al. Identification of MEDAG and SERPINE1 related to hypoxia in abdominal aortic aneurysm based on weighted gene coexpression network analysis. Front Physiol. 2022; Jul. 13:926508.24. Yang JD, Ma L, Zhu Z. SERPINE1 as a cancer-promoting gene in gastric adenocarcinoma: facilitates tumour cell proliferation, migration, and invasion by regulating EMT. J Chemother. 2019; Nov-Dec. 31(7-8):408–18.
Article25. Yen HC, Li SH, Majima HJ, Huang YH, Chen Cp, Liu CC, et al. Upregulation of antioxidant enzymes and coenzyme Q(10) in a human oral cancer cell line with acquired bleomycin resistance. Free Radic Res. 2011; Jun. 45(6):707–16.26. Cui J, Wang H, Zhang X, Sun X, Zhang J, Ma J. Exosomal miR-200c suppresses chemoresistance of docetaxel in tongue squamous cell carcinoma by suppressing TUBB3 and PPP2R1B. Aging (Albany NY). 2020; Apr. 12(8):6756–73.27. Bie Q, Song H, Chen X, Yang X, Shi S, Zhang L, et al. IL-17B/IL-17RB signaling cascade contributes to self-renewal and tumorigenesis of cancer stem cells by regulating Beclin-1 ubiquitination. Oncogene. 2021; Mar. 40(12):2200–16.
Article28. Yan T, Tan Y, Deng G, Sun Z, Liu B, Wang Y, et al. TGF-β induces GBM mesenchymal transition through upregulation of CLDN4 and nuclear translocation to activate TNF-α/NF-κB signal pathway. Cell Death Dis. 2022; Apr. 13(4):339.29. Maimela NR, Liu S, Zhang Y. Fates of CD8+T cells in tumor microenvironment. Comput Struct Biotechnol J. 2018; Nov. 17:1–13.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Signs and symptoms of nicotine withdrawal and adverse effects of nicotine patch
- Psychometric Tools Related to the Assessment of Nicotine Dependence and Withdrawal Symptoms
- Cigarette Smoking, Stage of Smoking Cessation, Nicotine Dependency, and Urine Nicotine Among Smoking Adults with Diabetes
- Experimental Studies on the Relation between Nicotine and Sexual Hormone: Part IV. The Antidotal Action of Luteohormone on Nicotine Toxicity during Anaphylaxis: C. The Effect of Nicotine with Luteohormone on Anaphylaxis
- Survivin, Possible Marker and Prognostic Factor in Oral Squamous Cell Carcinomas