Cardiovasc Prev Pharmacother.
2020 Apr;2(2):50-55. 10.36011/cpp.2020.2.e6.
From Traditional Statistical Methods to Machine and Deep Learning for Prediction Models
- Affiliations
-
- 1Department of Biostatistics, Wonju College of Medicine, Yonsei University, Wonju, Korea
- 2Department of Precision Medicine, Wonju College of Medicine, Yonsei University, Wonju, Korea
- KMID: 2536974
- DOI: http://doi.org/10.36011/cpp.2020.2.e6
Abstract
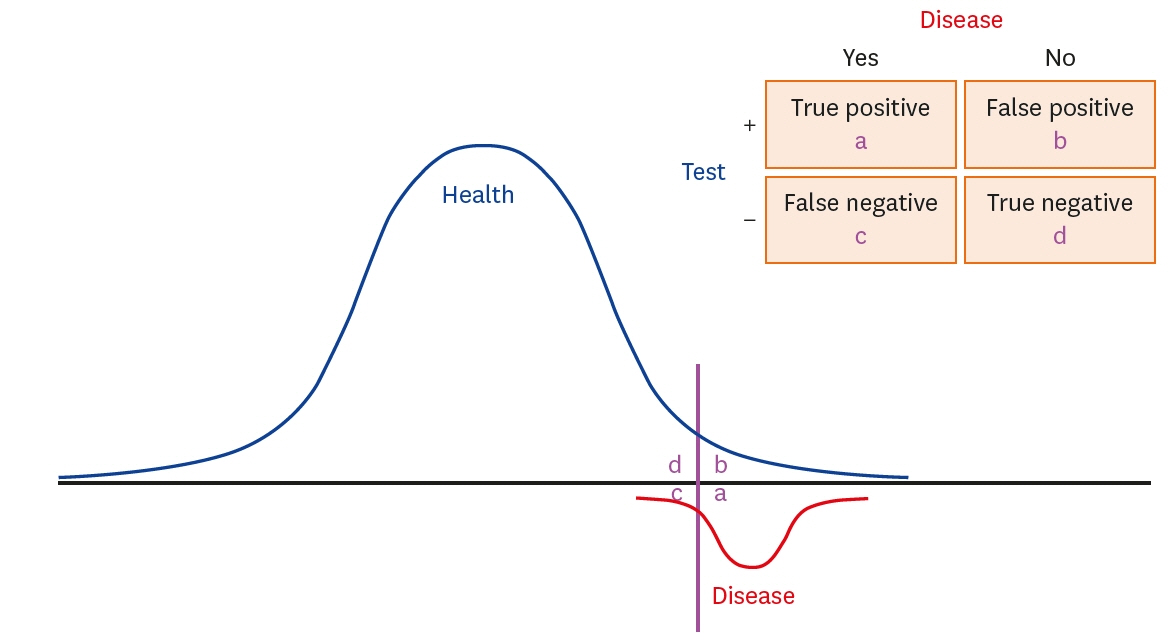
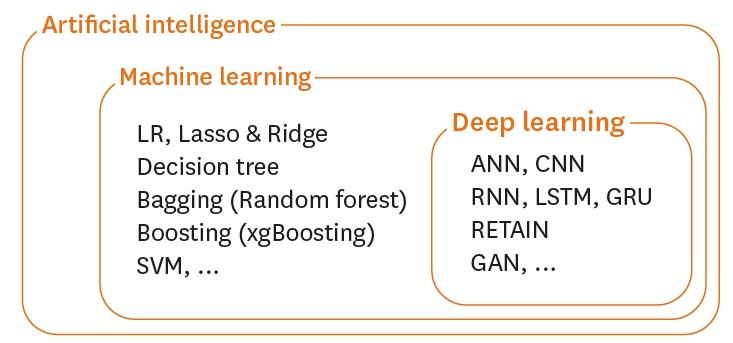
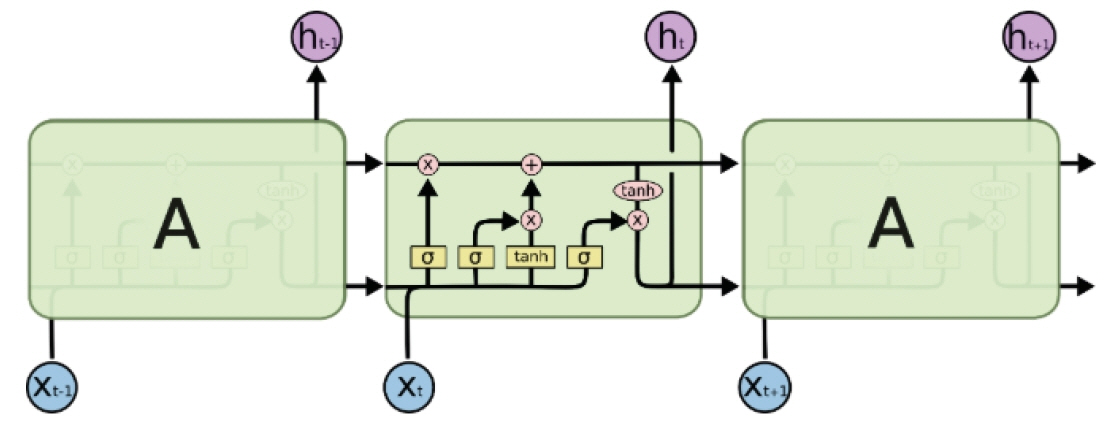
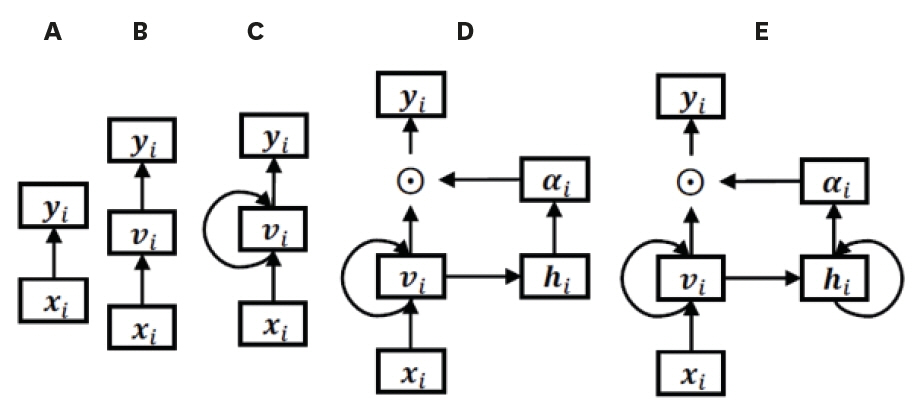
- Traditional statistical methods have low accuracy and predictability in the analysis of large amounts of data. In this method, non-linear models cannot be developed. Moreover, methods used to analyze data for a single time point exhibit lower performance than those used to analyze data for multiple time points, and the difference in performance increases as the amount of data increases. Using deep learning, it is possible to build a model that reflects all information on repeated measures. A recurrent neural network can be built to develop a predictive model using repeated measures. However, there are long-term dependencies and vanishing gradient problems. Meanwhile, long short-term memory method can be applied to solve problems with long-term dependency and vanishing gradient by assigning a fixed weight inside the cell state. Unlike traditional statistical methods, deep learning methods allow researchers to build non-linear models with high accuracy and predictability, using information from multiple time points. However, deep learning models cannot be interpreted; although, recently, many methods have been developed to do so by weighting time points and variables using attention algorithms, such as ReversE Time AttentIoN (RETAIN). In the future, deep learning methods, as well as traditional statistical methods, will become essential methods for big data analysis.
Keyword
Figure
Reference
-
1. Cho IJ, Sung JM, Chang HJ, Chung N, Kim HC. Incremental value of repeated risk factor measurements for cardiovascular disease prediction in middle-aged Korean adults: results from the NHIS-HEALS (National Health Insurance System-National Health Screening Cohort). Circ Cardiovasc Qual Outcomes. 2017; 10:e004197.
Article2. Goldstein BA, Navar AM, Pencina MJ, Ioannidis JP. Opportunities and challenges in developing risk prediction models with electronic health records data: a systematic review. J Am Med Inform Assoc. 2017; 24:198–208.
Article3. Paige E, Barrett J, Pennells L, Sweeting M, Willeit P, Di Angelantonio E, Gudnason V, Nordestgaard BG, Psaty BM, Goldbourt U, Best LG, Assmann G, Salonen JT, Nietert PJ, Verschuren WM, Brunner EJ, Kronmal RA, Salomaa V, Bakker SJ, Dagenais GR, Sato S, Jansson JH, Willeit J, Onat A, de la Cámara AG, Roussel R, Völzke H, Dankner R, Tipping RW, Meade TW, Donfrancesco C, Kuller LH, Peters A, Gallacher J, Kromhout D, Iso H, Knuiman M, Casiglia E, Kavousi M, Palmieri L, Sundström J, Davis BR, Njølstad I, Couper D, Danesh J, Thompson SG, Wood A. Use of repeated blood pressure and cholesterol measurements to improve cardiovascular disease risk prediction: an individual-participant-data metaanalysis. Am J Epidemiol. 2017; 186:899–907.
Article4. Zack CJ, Senecal C, Kinar Y, Metzger Y, Bar-Sinai Y, Widmer RJ, Lennon R, Singh M, Bell MR, Lerman A, Gulati R. Leveraging machine learning techniques to forecast patient prognosis after percutaneous coronary intervention. JACC Cardiovasc Interv. 2019; 12:1304–11.
Article5. Lee D, Yoo JK. The use of joint hierarchical generalized linear models: application to multivariate longitudinal data. Korean J Appl Stat. 2015; 28:335–42.
Article6. Ahn S. Deep learning architectures and applications. J Intell Inf Syst. 2016; 22:127–42.
Article7. Olah C. Understanding LSTM networks [Internet]. [place unknown]: colah's blog;2015. [cited 2020 Mar]. Available from: https://colah.github.io/posts/2015-08-Understanding-LSTMs/.8. Lipton ZC, Berkowitz J, Elkan C. A critical review of recurrent neural networks for sequence learning. arXiv 2015:1506.00019.9. Liu P, Qiu X, Huang X. Recurrent neural network for text classification with multi-task learning. arXiv 2016:1605.05101.10. Hochreiter S, Bengio Y, Frasconi P, Schmidhuber J. Gradient flow in recurrent nets: the difficulty of learning long-term dependencies. In : Kolen JF, Kremer SC, editors. A Field Guide to Dynamical Recurrent Neural Networks. New York, NY: IEEE Press;2001. p. 237–44.11. Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput. 1997; 9:1735–80.
Article12. Cho K, Van Merriënboer B, Gulcehre C, Bahdanau D, Bougares F, Schwenk H, Bengio Y. Learning phrase representations using RNN encoder-decoder for statistical machine translation. arXiv 2014:1406.1078.13. Choi E, Bahadori MT, Sun J, Kulas J, Schuetz A, Stewart W. Retain: an interpretable predictive model for healthcare using reverse time attention mechanism. arXiv 2016:1608.05745.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Machine Learning vs. Statistical Model for Prediction Modelling: Application in Medical Imaging Research
- Use of Machine Learning in Stroke Rehabilitation: A Narrative Review
- Artificial intelligence, machine learning, and deep learning in women’s health nursing
- Statistics and Deep Belief Network-Based Cardiovascular Risk Prediction
- Review of Statistical Methods for Evaluating the Performance of Survival or Other Time-to-Event Prediction Models (from Conventional to Deep Learning Approaches)