J Korean Neurosurg Soc.
2022 Sep;65(5):697-709. 10.3340/jkns.2021.0200.
Identification of Hub Genes in the Pathogenesis of Ischemic Stroke Based on Bioinformatics Analysis
- Affiliations
-
- 1Genetic Testing Center, The First Affiliated hospital of Dali University, Yunnan, China
- 2Clinical Colllege of Dali University, Yunnan, China
- KMID: 2533032
- DOI: http://doi.org/10.3340/jkns.2021.0200
Abstract
Objective
: The present study aimed to identify the function of ischemic stroke (IS) patients’ peripheral blood and its role in IS, explore the pathogenesis, and provide direction for clinical research progress by comprehensive bioinformatics analysis.
Methods
: Two datasets, including GSE58294 and GSE22255, were downloaded from Gene Expression Omnibus database. GEO2R was utilized to obtain differentially expressed genes (DEGs). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of DEGs were performed using the database annotation, visualization and integrated discovery database. The protein-protein interaction (PPI) network of DEGs was constructed by search tool of searching interactive gene and visualized by Cytoscape software, and then the Hub gene was identified by degree analysis. The microRNA (miRNA) and miRNA target genes closely related to the onset of stroke were obtained through the miRNA gene regulatory network.
Results
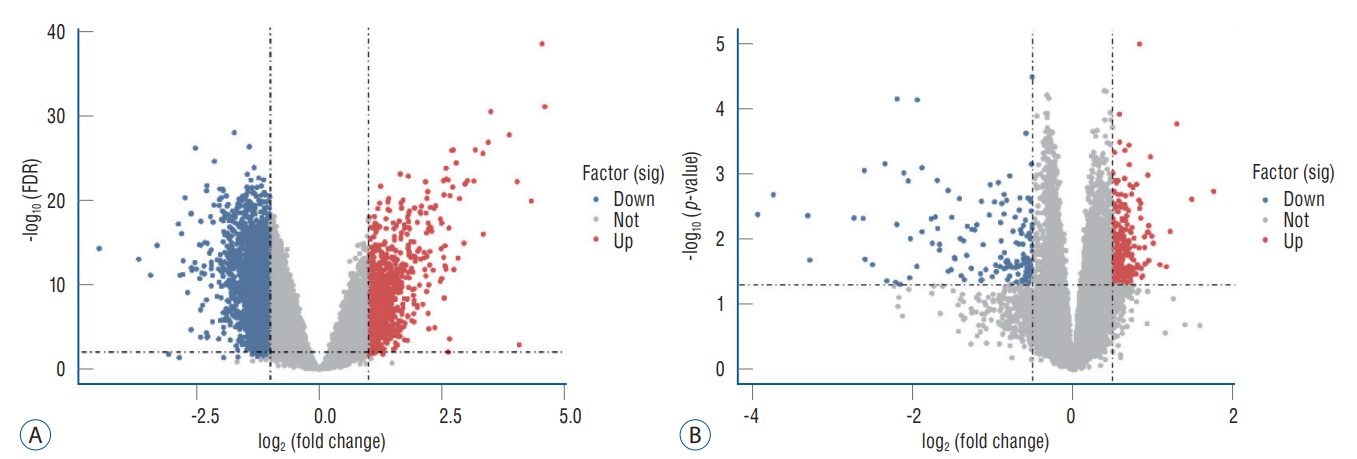
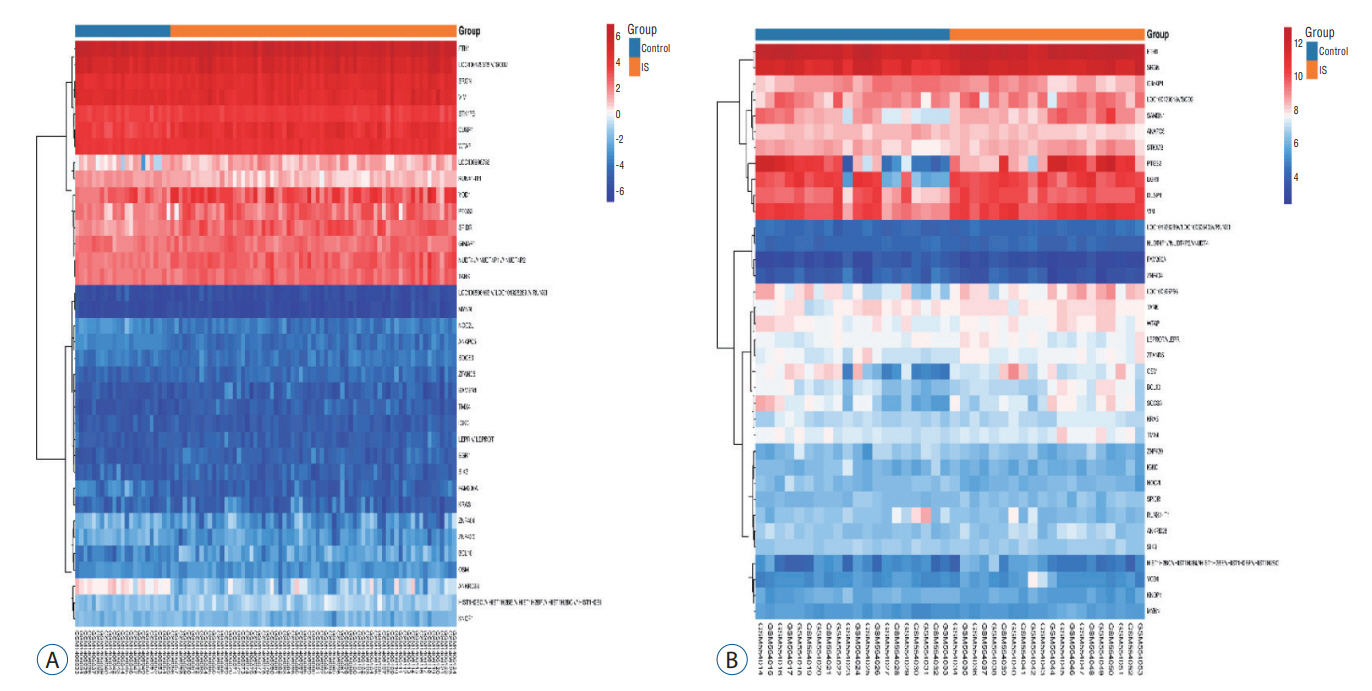
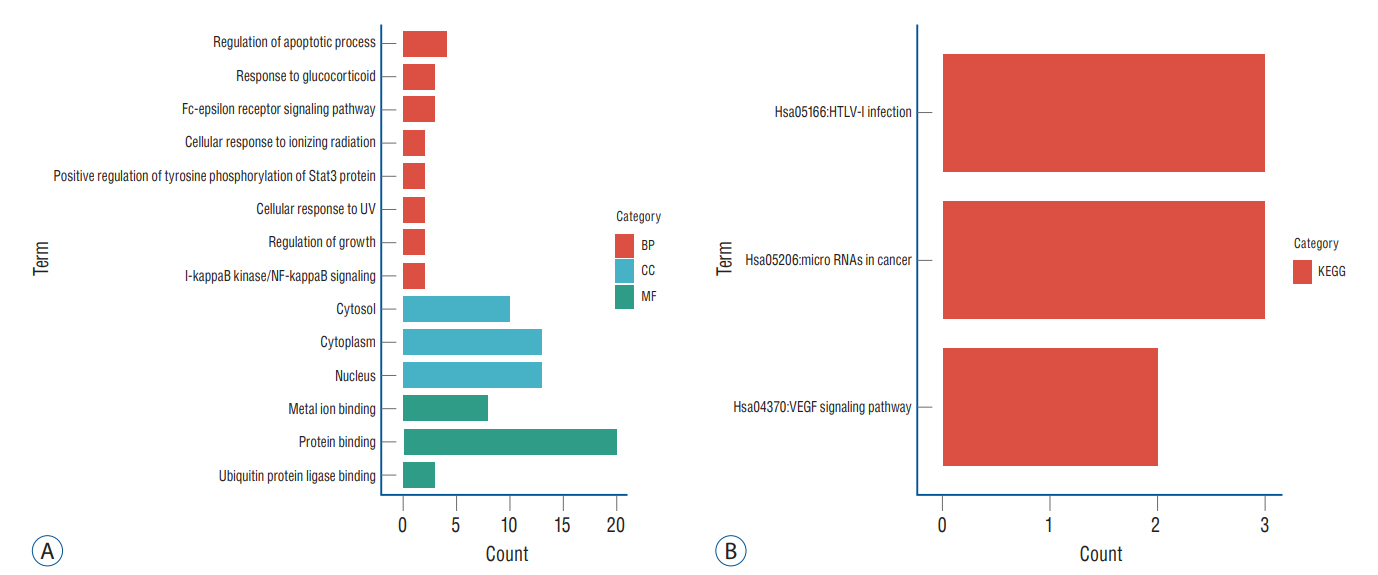
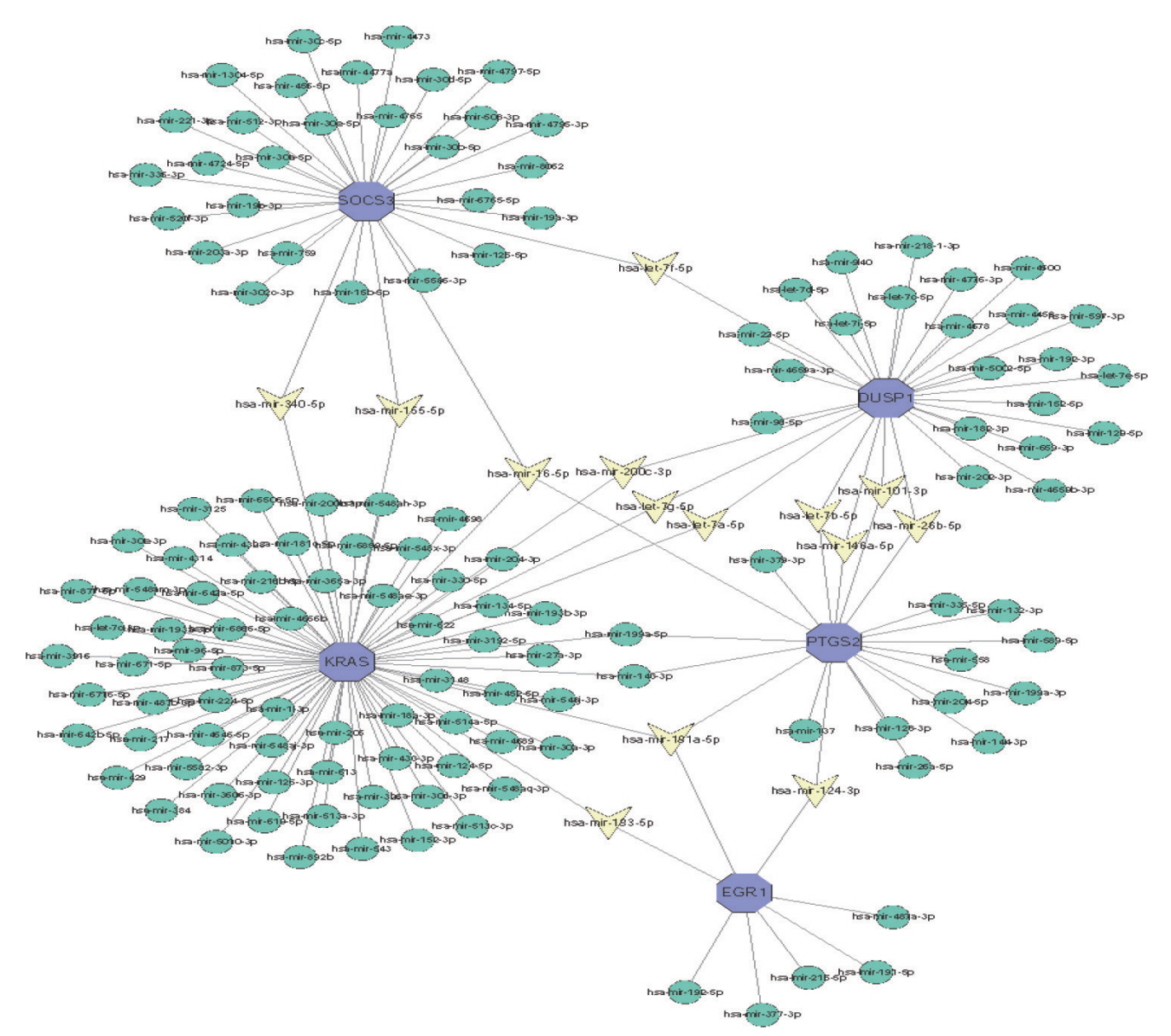
: In total, 36 DEGs, containing 27 up-regulated and nine down-regulated DEGs, were identified. GO functional analysis showed that these DEGs were involved in regulation of apoptotic process, cytoplasm, protein binding and other biological processes. KEGG enrichment analysis showed that these DEGs mediated signaling pathways, including HTLV-I infection and microRNAs in cancer. The results of PPI network and cytohubba showed that there was a relationship between DEGs, and five hub genes related to stroke were obtained : SOCS3, KRAS, PTGS2, EGR1, and DUSP1. Combined with the visualization of DEG-miRNAs, hsa-mir-16-5p, hsa-mir-181a-5p and hsa-mir-124-3p were predicted to be the key miRNAs in stroke, and three miRNAs were related to hub gene.
Conclusion
: Thirty-six DEGs, five Hub genes, and three miRNA were obtained from bioinformatics analysis of IS microarray data, which might provide potential targets for diagnosis and treatment of IS.
Figure
Reference
-
References
1. An YC, Tong SF, Wang D, Zhang Q, Su HL. MiR-16-5p targets TXNIP to regulate LPS-induced oxidative stress and apoptosis in cardiomyocytes. Chinese J of BMB. 36:934–944. 2020.2. Cao Y, Zhang D, Moon HG, Lee H, Haspel JA, Hu K, et al. MiR-15a/16 regulates apoptosis of lung epithelial cells after oxidative stress. Mol Med. 22:233–243. 2016.3. Chen G, Shen ZL, Wang L, Lv CY, Huang XE, Zhou RP. Hsa-miR-181a-5p expression and effects on cell proliferation in gastric cancer. Asian Pac J Cancer Prev. 14:3871–3875. 2013.4. Cheng ZH, Zhang YL, Huang W, Zhou PL, Luo LJ. Correlation of neurological function recovery with inflammatory factors in acute cerebral infarction patients. Hainan Med J. 30:2456–2458. 2019.5. Colaizzo D, Fofi L, Tiscia G, Guglielmi R, Cocomazzi N, Prencipe M, et al. The COX-2 G/C -765 polymorphism may modulate the occurrence of cerebrovascular ischemia. Blood Coagul Fibrinolysis. 17:93–96. 2006.6. Cui Z, Liang B, Li G, Li H. Expressions of miR-181a-5p and miR-126 in the serum of patients with inflammatory bowel disease and their correlation with intestinal flora. Intl J of DIGEST DIS. 41:33–37. 2021.7. Du Y, Chen X, Zhao S. Progress of mechanism of oxidative stress in acute ischemic stroke. Chinses J of Neuro Srg. 7:121–124. 2021.8. Ehlting C, Lai WS, Schaper F, Brenndörfer ED, Matthes RJ, Heinrich PC, et al. Regulation of suppressor of cytokine signaling 3 (SOCS3) mRNA stability by TNF-alpha involves activation of the MKK6/p38MAPK/MK2 cascade. J Immunol. 178:2813–2826. 2007.9. Fantini D, Vascotto C, Deganuto M, Bivi N, Gustincich S, Marcon G, et al. APE1/Ref-1 regulates PTEN expression mediated by Egr-1. Free Radic Res. 42:20–29. 2008.10. Feng WR, Xu Y, Yu WK, Wu T, Li J. Relationship between serum miR124 and miR-134 expression and severity of disease and inflammatory response in patients with acute ischemic stroke. Prog Mod Bod. 21:1751–1754. 2021.11. He M, Yu H, Yang W, Yang Y, Su W. Mechanism of miR-124-3p targeting Rab11a to alleviate oxidative stress and inflammatory response induced by traumatic brain injury. Intl J of Lab Med. 42:1195–1199. 2021.12. Hong F, Nguyen VA, Gao B. Tumor necrosis factor alpha attenuates interferon alpha signaling in the liver: involvement of SOCS3 and SHP2 and implication in resistance to interferon therapy. FASEB J. 15:1595–1597. 2001.13. Huang D, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4:44–57. 2009.14. Institute for Health Metrics and Evaluation : Global Health Data Exchange. GBD Results Tool [DB/OL]. Available at : http://ghdx.healthdata.org/gbd-results-tool.15. Jiang M, Wang H, Jin M, Yang X, Ji H, Jiang Y, et al. Exosomes from miR-30d-5p-ADSCs reverse acute ischemic stroke-induced, autophagy-mediated brain injury by promoting M2 microglial/macrophage polarization. Cell Physiol Biochem. 47:864–878. 2018.16. Kim AS, Cahill E, Cheng NT. Global stroke belt: geographic variation in stroke burden worldwide. Stroke. 46:3564–3570. 2015.17. Kohsaka S, Volcik KA, Folsom AR, Wu KK, Ballantyne CM, Willerson JT, et al. Increased risk of incident stroke associated with the cyclooxygenase 2 (COX-2) G-765C polymorphism in african-americans: the atherosclerosis risk in communities study. Atherosclerosis. 196:926–930. 2008.18. Korhan P, Erdal E, Atabey N. MiR-181a-5p is downregulated in hepatocellular carcinoma and suppresses motility, invasion and branchin-gmorphogenesis by directly targeting C-Met. Biochem Biophys Res Commun. 450:1304–1312. 2014.19. Kumar A, Misra S, Sagar R, Kumar P, Yadav AK, Talwar P, et al. Relationship between factor V Leiden gene variant and risk of ischemic stroke: a case-control study. Ann Indian Acad Neurol. 20:284–288. 2017.20. Li P, Li F, Gao F, Zhang C. The expressions of Caveolin-1 and miR-199 a-5 p in peripheral blood of is-chemic stroke patients and in brain and spleen tissues of rat models. Chinese J of Gero. 39:1558–1562. 2019.21. Liu K, Xie F, Gao A, Zhang R, Zhang L, Xiao Z, et al. SOX2 regulates multiple malignant processes of breast cancer development through the SOX2/miR-181a-5p, miR-30e-5p/TUSC3 axis. Mol Cancer. 16:62. 2017.22. Meng Q, Liu Y, Huo X, Sun H, Wang Y, Bu F. MicroRNA-221-3p contributes to cardiomyocyte injury in H2O2-treated H9c2 cells and a rat model of myocardial ischemia-reperfusion by targeting p57. Int J Mol Med. 42:589–596. 2018.23. Moran A, Gu D, Zhao D, Coxson P, Wang YC, Chen CS, et al. Future cardiovascular disease in China: markov model and risk factor scenario projections from the coronary heart disease policy model-China. Circ Cardiovasc Qual Outcomes. 3:243–252. 2010.24. O’Sullivan LA, Liongue C, Lewis RS, Stephenson SE, Ward AC. Cytokine receptor signaling through the jak-stat-socs pathway in disease. Mol Immunol. 44:2497–2506. 2007.25. Pagel JI, Deindl E. Early growth response 1--a transcription factor in the crossfire of signal transduction cascades. Indian J Biochem Biophys. 48:226–235. 2011.26. Pan Y, Hong Y, Zhang QY, Kong LD. Impaired hypothalamic insulin signaling in CUMS rats: restored by icariin and fluoxetine through inhibiting CRF system. Psychoneuroendocrinology. 38:122–134. 2013.27. Sarkar IN. Biomedical informatics and translational medicine. J Transl Med. 8:22. 2010.28. Shin IS, Kim JM, Kim KL, Jang SY, Jeon ES, Choi SH, et al. Early growth response factor-1 is associated with intraluminal thrombus formation in human abdominal aortic aneurysm. J Am Coll Cardiol. 53:792–799. 2009.29. Wang L, Liu J, Yang Y, Peng B, Wang YL. The prevention and treatment of stroke still face huge challenges-brief report on stroke prevention and treatment in China, 2018. Chin Circ J. 34:105–119. 2019.30. Wang NP, Pang XF, Zhang LH, Tootle S, Harmouche S, Zhao ZQ. Attenuation of inflammatory response and reduction in infarct size by postconditioning are associated with downregulation of early growth response 1 during reperfusion in rat heart. Shock. 41:346–354. 2014.31. Wang W, Jiang B, Sun H, Ru X, Sun D, Wang L, et al. Prevalence, incidence, and mortality of stroke in China: results from a nationwide population-based survey of 480 687 adults. Circulation. 135:759–771. 2017.32. Wu KK, Liou JY, Cieslik K. Transcriptional control of COX-2 via C/EBPbeta. Arterioscler Thromb Vasc Biol. 25:679–685. 2005.33. Yi JH, Park SW, Kapadia R, Vemuganti R. Role of transcription factors in mediating post-ischemic cerebral inflammation and brain damage. Neurochem Int. 50:1014–1027.34. Zhang WN, Wang L, Wang Q, Luo X, Fang DF, Chen Y, et al. CUEDC2 (CUE domain-containing 2) and SOCS3 (suppressors of cytokine signaling 3) cooperate to negatively regulate Janus kinase 1/signal transducers and activators of transcription 3 signaling. J Biol Chem. 287:382–392. 2012.35. Zhang Y, Wang X, Xu X, Chen R, Kan H. Stock volatility and stroke mortality in a Chinese population. J Cardiovasc Med (Hagerstown). 14:617–621. 2013.36. Zhnag S, Wang F, Li H, Wen Y. Changes and significance of plasma miR-146a and miR-21 expression in patients with acute ischemic stroke after thrombolysis. Shangdong Med J. 59:61–63. 2019.37. Zhou J, Chen L, Chen B, Huang S, Zeng C, Wu H, et al. Increased serum exosomal miR-134 expression in the acute ischemic stroke patients. BMC Neurol. 18:198. 2018.38. Zhu Z, Fu Y, Tian D, Sun N, Han W, Chang G, et al. Combination of the immune modulator fingolimod with alteplase in acute ischemic stroke: a pilot trial. Circulation. 132:1104–1112. 2015.39. Zuo H, Yue X, Gui Y, Ren R, Wang Z, Zhao J, et al. Regulatory effect of miR-145-5p targeting FGF5 on apoptosis and oxidative stress induced by ischemia/reperfusion injury. Acta Univ Med Anhui. 54:887–893. 2019.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Identification of multiple hub genes in acute kidney injury after kidney transplantation by bioinformatics analysis
- Exploring molecular mechanisms underlying the pathophysiological association between knee osteoarthritis and sarcopenia
- A network-biology approach for identification of key genes and pathways involved in malignant peritoneal mesothelioma
- Identification of Differentially-Methylated Genes and Pathways in Patients with Delayed Cerebral Ischemia Following Subarachnoid Hemorrhage
- Potential biomarkers and signaling pathways associated with the pathogenesis of primary salivary gland carcinoma: a bioinformatics study