J Korean Neurosurg Soc.
2022 Jan;65(1):4-12. 10.3340/jkns.2021.0035.
Identification of Differentially-Methylated Genes and Pathways in Patients with Delayed Cerebral Ischemia Following Subarachnoid Hemorrhage
- Affiliations
-
- 1Institute of New Frontier Stroke Research, Hallym University College of Medicine, Chuncheon, Korea
- 2Department of Neurosurgery, Hallym University College of Medicine, Chuncheon, Korea
- 3Department of Microbiology, College of Science & Technology, Dankook University, Cheonan, Korea
- 4Genetic and Research Inc., Chuncheon, Korea
- KMID: 2523908
- DOI: http://doi.org/10.3340/jkns.2021.0035
Abstract
Objective
: We reported the differentially methylated genes in patients with subarachnoid hemorrhage (SAH) using bioinformatics analyses to explore the biological characteristics of the development of delayed cerebral ischemia (DCI).
Methods
: DNA methylation profiles obtained from 40 SAH patients from an epigenome-wide association study were analyzed. Functional enrichment analysis, protein-protein interaction (PPI) network, and module analyses were carried out.
Results
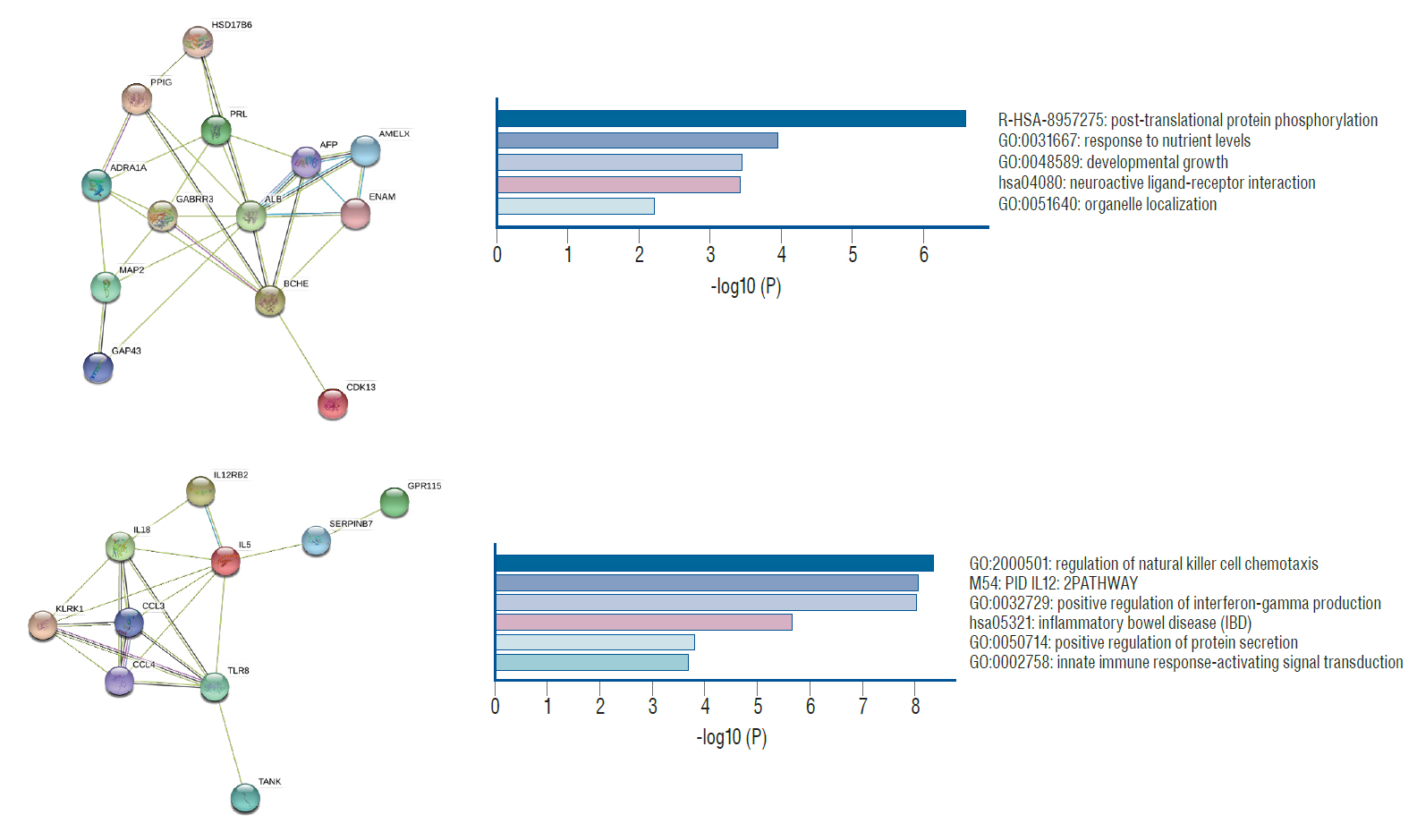
: A total of 13 patients (32.5%) experienced DCI during the follow-up. In total, we categorized the genes into the two groups of hypermethylation (n=910) and hypomethylation (n=870). The hypermethylated genes referred to biological processes of organic cyclic compound biosynthesis, nucleobase-containing compound biosynthesis, heterocycle biosynthesis, aromatic compound biosynthesis and cellular nitrogen compound biosynthesis. The hypomethylated genes referred to biological processes of carbohydrate metabolism, the regulation of cell size, and the detection of a stimulus, and molecular functions of amylase activity, and hydrolase activity. Based on PPI network and module analysis, three hypermethylation modules were mainly associated with antigen-processing, Golgi-to-ER retrograde transport, and G alpha (i) signaling events, and two hypomethylation modules were associated with post-translational protein phosphorylation and the regulation of natural killer cell chemotaxis. VHL, KIF3A, KIFAP3, RACGAP1, and OPRM1 were identified as hub genes for hypermethylation, and ALB and IL5 as hub genes for hypomethylation.
Conclusion
: This study provided novel insights into DCI pathogenesis following SAH. Differently methylated hub genes can be useful biomarkers for the accurate DCI diagnosis.
Keyword
Figure
Reference
-
References
1. Babu MS, Kaul S, Dadheech S, Rajeshwar K, Jyothy A, Munshi A. Serum albumin levels in ischemic stroke and its subtypes: correlation with clinical outcome. Nutrition. 29:872–875. 2013.
Article2. Chidambaran V, Zhang X, Martin LJ, Ding L, Weirauch MT, Geisler K, et al. DNA methylation at the mu-1 opioid receptor gene (OPRM1) promoter predicts preoperative, acute, and chronic postsurgical pain after spine fusion. Pharmgenomics Pers Med. 10:157–168. 2017.3. Dorsch NW, King MT. A review of cerebral vasospasm in aneurysmal subarachnoid haemorrhage part I: incidence and effects. J Clin Neurosci. 1:19–26. 1994.
Article4. Duan W, Pan Y, Wang C, Wang Y, Zhao X, Wang Y, et al. Risk factors and clinical impact of delayed cerebral ischemia after aneurysmal subarachnoid hemorrhage: analysis from the China National Stroke Registry. Neuroepidemiology. 50:128–136. 2018.
Article5. Endres M, Meisel A, Biniszkiewicz D, Namura S, Prass K, Ruscher K, et al. DNA methyltransferase contributes to delayed ischemic brain injury. J Neurosci. 20:3175–3181. 2000.
Article6. Francoeur CL, Mayer SA. Management of delayed cerebral ischemia after subarachnoid hemorrhage. Crit Care. 20:277. 2016.
Article7. Guinot A, Lehmann H, Wild PJ, Frew IJ. Combined deletion of Vhl, Trp53 and Kif3a causes cystic and neoplastic renal lesions. J Pathol. 239:365–373. 2016.
Article8. Hamzei Moqaddam A, Ahmadi Musavi SM, Khademizadeh K. Relationship of opium dependency and stroke. Addict Health. 1:6–10. 2009.9. Herman JG, Latif F, Weng Y, Lerman MI, Zbar B, Liu S, et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci U S A. 91:9700–9704. 1994.
Article10. Ieda M, Miyaoka T, Kawano K, Wake R, Inagaki T, Horiguchi J. May salivary alpha-amylase level be a useful tool for assessment of the severity of schizophrenia and evaluation of therapy? A case report. Case Rep Psychiatry. 2012:747104. 2012.
Article11. Kim BJ, Kim Y, Hong EP, Jeon JP, Yang JS, Choi HJ, et al. Correlation between altered DNA methylation of intergenic regions of ITPR3 and development of delayed cerebral ischemia in patients with subarachnoid hemorrhage. World Neurosurg. 130:e449–e456. 2019.
Article12. Kim BJ, Kim Y, Kim SE, Jeon JP. Study of correlation between Hp α1 expression of haptoglobin 2-1 and clinical course in aneurysmal subarachnoid hemorrhage. World Neurosurg. 117:e221–e227. 2018.
Article13. Kim BJ, Kim Y, Youn DH, Park JJ, Rhim JK, Kim HC, et al. Genome-wide blood DNA methylation analysis in patients with delayed cerebral ischemia after subarachnoid hemorrhage. Sci Rep. 10:11419. 2020.
Article14. Kim H, Crago E, Kim M, Sherwood P, Conley Y, Poloyac S, et al. Cerebral vasospasm after sub-arachnoid hemorrhage as a clinical predictor and phenotype for genetic association study. Int J Stroke. 8:620–625. 2013.
Article15. Knutsson A, Björkbacka H, Dunér P, Engström G, Binder CJ, Nilsson AH, et al. Associations of interleukin-5 with plaque development and cardiovascular events. JACC Basic Transl Sci. 4:891–902. 2019.
Article16. Kovacic MB, Myers JM, Wang N, Martin LJ, Lindsey M, Ericksen MB, et al. Identification of KIF3A as a novel candidate gene for childhood asthma using RNA expression and population allelic frequencies differences. PLoS One. 6:e23714. 2011.
Article17. Kurmi K, Haigis MC. Nitrogen metabolism in cancer and immunity. Trends Cell Biol. 30:408–424. 2020.
Article18. Kyadav K, Chaudhary HR, Gupta RC, Jain R, Yadav SR, Sharma S, et al. Clinical profile and outcome of stroke in relation to glycaemic status of patients. J Indian Med Assoc. 102:138–150. 138-139, 142, 150. 2004.19. Lad SP, Hegen H, Gupta G, Deisenhammer F, Steinberg GK. Proteomic biomarker discovery in cerebrospinal fluid for cerebral vasospasm following subarachnoid hemorrhage. J Stroke Cerebrovasc Dis. 21:30–41. 2012.
Article20. Lei L, Mason S, Liu D, Huang Y, Marks C, Hickey R, et al. Hypoxiainducible factor-dependent degeneration, failure, and malignant transformation of the heart in the absence of the von Hippel-Lindau protein. Mol Cell Biol. 28:3790–3803. 2008.
Article21. Li X, Lin S, Chen X, Huang W, Li Q, Zhang H, et al. The Prognostic value of serum cytokines in patients with acute ischemic stroke. Aging Dis. 10:544–556. 2019.
Article22. Liu D, Zempleni J. Transcriptional regulation of the albumin gene depends on the removal of histone methylation marks by the FAD-dependent monoamine oxidase lysine-specific demethylase 1 in HepG2 human hepatocarcinoma cells. J Nutr. 144:997–1001. 2014.
Article23. Liu Y, Gu HY, Zhu J, Niu YM, Zhang C, Guo GL. Identification of Hub genes and key pathways associated with bipolar disorder based on weighted gene co-expression network analysis. Front Physiol. 10:1081. 2019.
Article24. Martynov IuS, Shuvakhina NA. Carbohydrate metabolism disorders in stroke. Zh Nevropatol Psikhiatr Im S S Korsakova. 79:1281–1288. 1979.25. McRonald FE, Morris MR, Gentle D, Winchester L, Baban D, Ragoussis J, et al. CpG methylation profiling in VHL related and VHL unrelated renal cell carcinoma. Mol Cancer. 8:31. 2009.
Article26. Nikkola E, Laiwalla A, Ko A, Alvarez M, Connolly M, Ooi YC, et al. Remote ischemic conditioning alters methylation and expression of cell cycle genes in aneurysmal subarachnoid hemorrhage. Stroke. 46:2445–2451. 2015.
Article27. Ott MO, Sperling L, Cassio D, Levilliers J, Sala-Trepat J, Weiss MC. Undermethylation at the 5' end of the albumin gene is necessary but not sufficient for albumin production by rat hepatoma cells in culture. Cell. 30:825–833. 1982.
Article28. Pascual JM, Carceller F, Roda JM, Cerdán S. Glutamate, glutamine, and GABA as substrates for the neuronal and glial compartments after focal cerebral ischemia in rats. Stroke. 29:1048–1056. discussion 1056-1057. 1998.
Article29. Peyravian N, Dikici E, Deo S, Toborek M, Daunert S. Opioid antagonists as potential therapeutics for ischemic stroke. Prog Neurobiol. 182:101679. 2019.
Article30. Qureshi WT, O'Neal WT, Khodneva Y, Judd S, Safford MM, Muntner P, et al. Association between opioid use and atrial fibrillation : the reasons for geographic and racial differences in stroke (REGARDS) study. JAMA Intern Med. 175:1058–1060. 2015.
Article31. Rowland MJ, Hadjipavlou G, Kelly M, Westbrook J, Pattinson KT. Delayed cerebral ischaemia after subarachnoid haemorrhage: looking beyond vasospasm. Br J Anaesth. 109:315–329. 2012.
Article32. Samuelsson C, Howells T, Kumlien E, Enblad P, Hillered L, Ronne-Engström E. Relationship between intracranial hemodynamics and microdialysis markers of energy metabolism and glutamate-glutamine turnover in patients with subarachnoid hemorrhage. Clinical article. J Neurosurg. 111:910–915. 2009.
Article33. Sang L, Wang XM, Xu DY, Zhao WJ. Bioinformatics analysis of aberrantly methylated-differentially expressed genes and pathways in hepatocellular carcinoma. World J Gastroenterol. 24:2605–2616. 2018.
Article34. Stamatovic SM, Phillips CM, Martinez-Revollar G, Keep RF, Andjelkovic AV. Involvement of epigenetic mechanisms and non-coding RNAs in blood-brain barrier and neurovascular unit injury and recovery after stroke. Front Neurosci. 13:864. 2019.
Article35. Starke RM, Kim GH, Komotar RJ, Hickman ZL, Black EM, Rosales MB, et al. Endothelial nitric oxide synthase gene single-nucleotide polymorphism predicts cerebral vasospasm after aneurysmal subarachnoid hemorrhage. J Cereb Blood Flow Metab. 28:1204–1211. 2008.
Article36. Suarez JI, Martin RH, Calvillo E, Dillon C, Bershad EM, Macdonald RL, et al. The albumin in subarachnoid hemorrhage (ALISAH) multicenter pilot clinical trial: safety and neurologic outcomes. Stroke. 43:683–690. 2012.
Article37. Zaina S, Heyn H, Carmona FJ, Varol N, Sayols S, Condom E, et al. DNA methylation map of human atherosclerosis. Circ Cardiovasc Genet. 7:692–700. 2014.
Article38. Zhao W, Lei T, Li H, Sun D, Mo X, Wang Z, et al. Macrophage-specific overexpression of interleukin-5 attenuates atherosclerosis in LDL receptor-deficient mice. Gene Ther. 22:645–652. 2015.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Relation between Thromboembolism and Delayed Ischemic Neurological Deficits in Aneurysmal Subarachnoid Hemorrhage
- Reliability of Transcranial Doppler Examination in the Diagnosis of Delayed Ischemia after Subarachnoid Hemorrhage
- Delayed Ischemic Deficit following Spontaneous Subarachnoid Homorrhage
- Two Cases of Subarachnoid Hemorrhage from Spontaneous Anterior Cerebral Artery Dissection : A Case of Simultaneous Hemorrhage and Ischemia Without Aneurysmal Formation and Another Case of Hemorrhage with Aneurysmal Formation
- Hemodynamics of Poor-grade Subarachnoid Hemorrhage