Int J Stem Cells.
2022 Aug;15(3):301-310. 10.15283/ijsc21035.
YBX1 Promotes the Inclusion of RUNX2 Alternative Exon 5 in Dental Pulp Stem Cells
- Affiliations
-
- 1The State Key Laboratory Breeding Base of Basic Science of Stomatology (Hubei-MOST) & Key Laboratory of Oral Biomedicine Ministry of Education, School & Hospital of Stomatology, Wuhan University, Wuhan, China
- 2Department of Orthodontics, Stomatological Hospital of Xiamen Medical College, Xiamen, China
- 3Department of Stomatology, Renmin Hospital of Wuhan University, Wuhan University, Wuhan, China
- 4Department of Endodontics, School & Hospital of Stomatology, Wuhan University, Wuhan, China
- KMID: 2532404
- DOI: http://doi.org/10.15283/ijsc21035
Abstract
- Background and Objectives
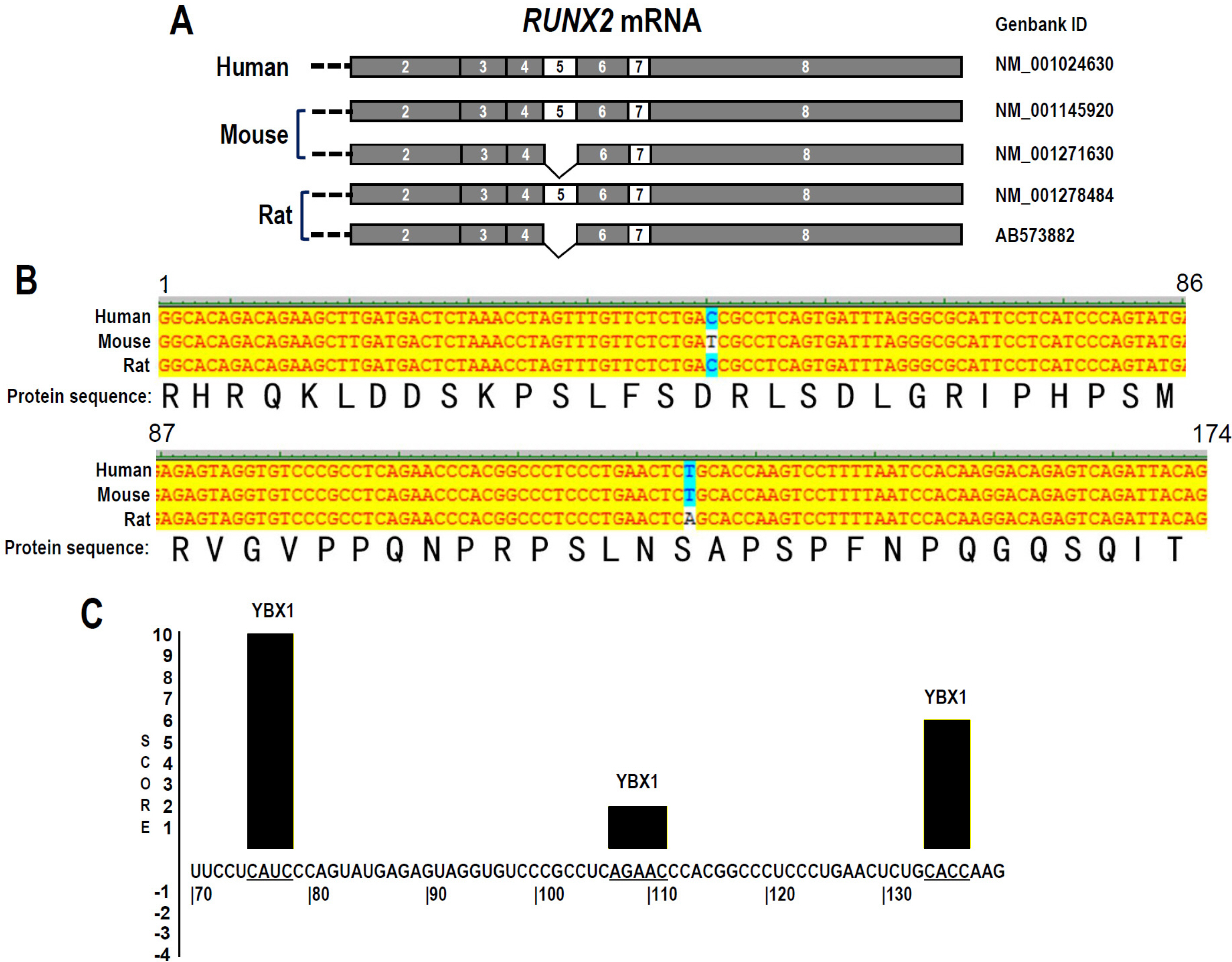
RUNX2 plays an essential role during the odontoblast differentiation of dental pulp stem cells (DPSCs). RUNX2 Exon 5 is an alternative exon and essential for RUNX2 transcriptional activity. This study aimed to investigate the regulatory mechanisms of RUNX2 exon 5 alternative splicing in human DPSCs.
Methods and Results
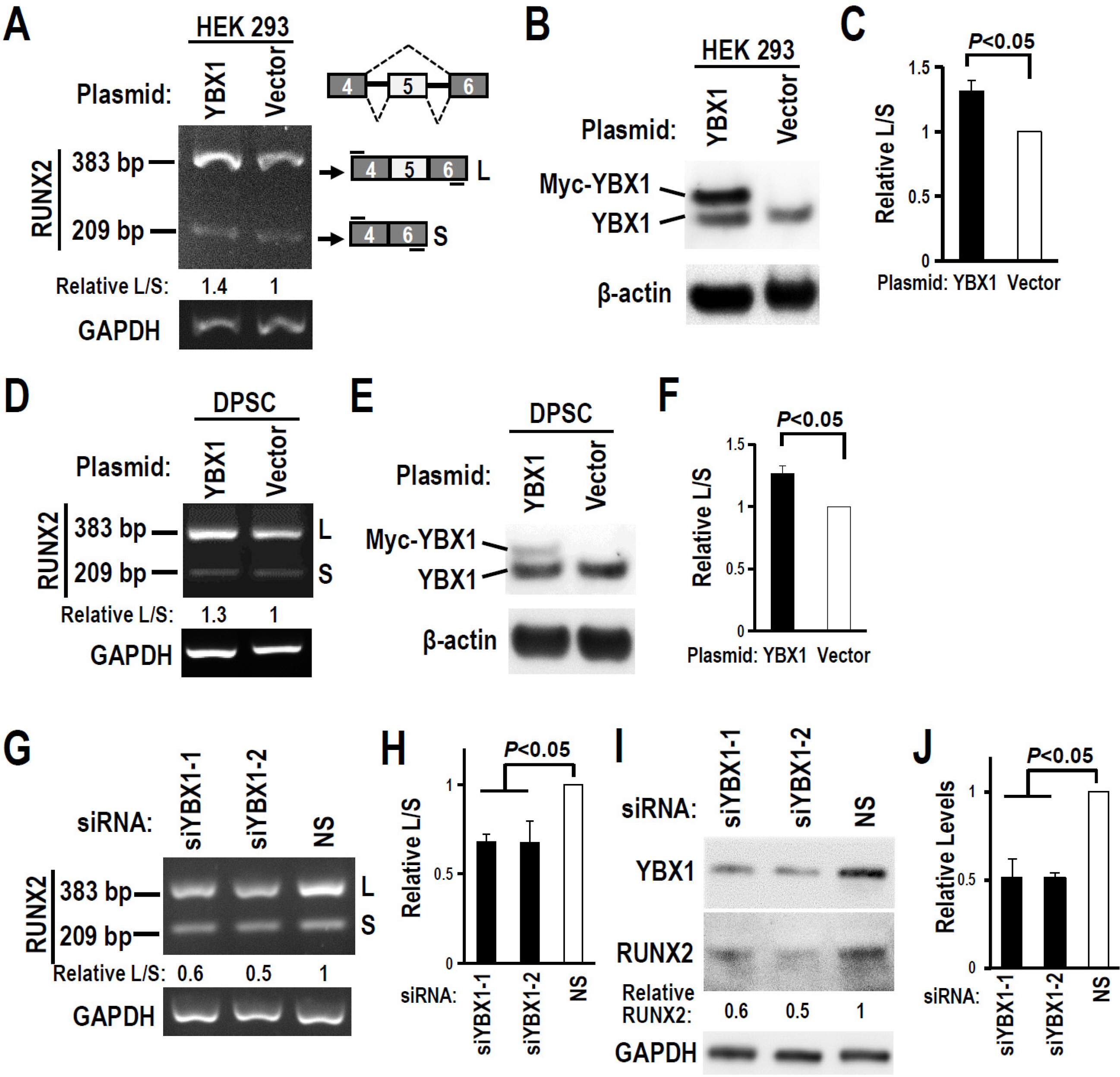
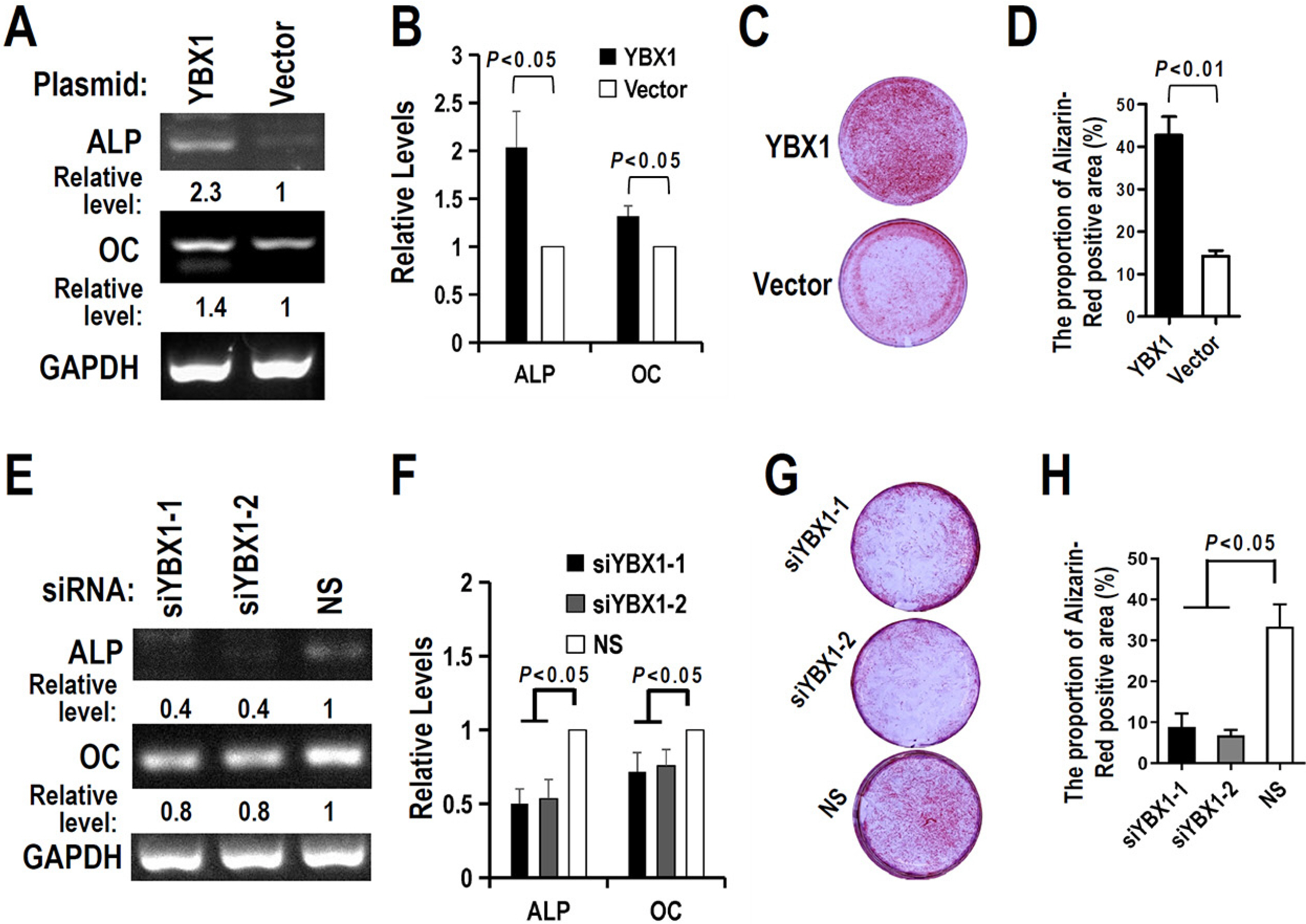
The regulatory motifs of RUNX2 exon 5 were analyzed using the online SpliceAid program. The alternative splicing of RUNX2 exon 5 in DPSCs during mineralization-induced differentiation was analyzed by RT-PCR. To explore the effect of splicing factor YBX1 on exon 5 alternative splicing, gaining or losing function of YBX1 was performed by transfection of YBX1 overexpression plasmid or anti-YBX1 siRNA in DPSCs. Human RUNX2 exon 5 is evolutionarily conserved and alternatively spliced in DPSCs. There are three potential YBX1 binding motifs in RUNX2 exon 5. The inclusion of RUNX2 exon 5 and YBX1 expression level increased significantly during mineralization-induced differentiation in DPSCs. Overexpression of YBX1 significantly increased the inclusion of RUNX2 exon 5 in DPSCs. In contrast, silence of YBX1 significantly reduced the inclusion of exon 5 and the corresponding RUNX2 protein expression level. Knockdown of YBX1 reduced the expression of alkaline phosphatase (ALP) and osteocalcin (OC) and the mineralization ability of DPSCs, while overexpression of YBX1 increased the expression of ALP and OC and the mineralization ability of DPSCs.
Conclusions
Human RUNX2 exon 5 is conserved evolutionarily and alternatively spliced in DPSCs. Splicing factor YBX1 promotes the inclusion of RUNX2 exon 5 and improves the mineralization ability of DPSCs.
Keyword
Figure
Reference
-
References
1. Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. 1997; Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 89:755–764. DOI: 10.1016/S0092-8674(00)80258-5. PMID: 9182763. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0030684749&origin=inward.2. Li S, Kong H, Yao N, Yu Q, Wang P, Lin Y, Wang J, Kuang R, Zhao X, Xu J, Zhu Q, Ni L. 2011; The role of runt-related transcription factor 2 (Runx2) in the late stage of odontoblast differentiation and dentin formation. Biochem Biophys Res Commun. 410:698–704. DOI: 10.1016/j.bbrc.2011.06.065. PMID: 21703228. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=79960031652&origin=inward.3. Cohen MM Jr. 2013; Biology of RUNX2 and cleidocranial dysplasia. J Craniofac Surg. 24:130–133. DOI: 10.1097/SCS.0b013e3182636b7e. PMID: 23348269. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84873289126&origin=inward.4. Levanon D, Groner Y. 2004; Structure and regulated expression of mammalian RUNX genes. Oncogene. 23:4211–4219. DOI: 10.1038/sj.onc.1207670. PMID: 15156175. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=2942512933&origin=inward.5. Vimalraj S, Arumugam B, Miranda PJ, Selvamurugan N. 2015; Runx2: structure, function, and phosphorylation in osteoblast differentiation. Int J Biol Macromol. 78:202–208. DOI: 10.1016/j.ijbiomac.2015.04.008. PMID: 25881954. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84928326539&origin=inward.6. Makita N, Suzuki M, Asami S, Takahata R, Kohzaki D, Kobayashi S, Hakamazuka T, Hozumi N. 2008; Two of four alternatively spliced isoforms of RUNX2 control osteocalcin gene expression in human osteoblast cells. Gene. 413:8–17. DOI: 10.1016/j.gene.2007.12.025. PMID: 18321663. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=41249084399&origin=inward.7. Ge J, Guo S, Fu Y, Zhou P, Zhang P, Du Y, Li M, Cheng J, Jiang H. 2015; Dental follicle cells participate in tooth eruption via the RUNX2-MiR-31-SATB2 loop. J Dent Res. 94:936–944. DOI: 10.1177/0022034515578908. PMID: 25818585. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84935126536&origin=inward.8. Narayanan A, Srinaath N, Rohini M, Selvamurugan N. 2019; Regulation of Runx2 by MicroRNAs in osteoblast differen-tiation. Life Sci. 232:116676. DOI: 10.1016/j.lfs.2019.116676. PMID: 31340165. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85069653667&origin=inward.9. Kim HJ, Kim WJ, Ryoo HM. 2020; Post-translational regulations of transcriptional activity of RUNX2. Mol Cells. 43:160–167. DOI: 10.14348/molcells.2019.0247. PMID: 31878768. PMCID: PMC7057842. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85081157528&origin=inward.10. Li YL, Xiao ZS. 2007; Advances in Runx2 regulation and its isoforms. Med Hypotheses. 68:169–175. DOI: 10.1016/j.mehy.2006.06.006. PMID: 16901655. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=33751103183&origin=inward.11. D'Souza RN, Aberg T, Gaikwad J, Cavender A, Owen M, Karsenty G, Thesleff I. 1999; Cbfa1 is required for epithelial-mesenchymal interactions regulating tooth development in mice. Development. 126:2911–2920. DOI: 10.1242/dev.126.13.2911. PMID: 10357935.12. Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. 2000; Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci U S A. 97:13625–13630. DOI: 10.1073/pnas.240309797. PMID: 11087820. PMCID: PMC17626. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0034610376&origin=inward.13. Kwon E, Todorova K, Wang J, Horos R, Lee KK, Neel VA, Negri GL, Sorensen PH, Lee SW, Hentze MW, Mandinova A. 2018; The RNA-binding protein YBX1 regulates epidermal progenitors at a posttranscriptional level. Nat Commun. 9:1734. DOI: 10.1038/s41467-018-04092-0. PMID: 29712925. PMCID: PMC5928080. PMID: ae77c37590464e83bc58037121dc3a32. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85046367125&origin=inward.14. Jia R, Liu X, Tao M, Kruhlak M, Guo M, Meyers C, Baker CC, Zheng ZM. 2009; Control of the papillomavirus early-to-late switch by differentially expressed SRp20. J Virol. 83:167–180. DOI: 10.1128/JVI.01719-08. PMID: 18945760. PMCID: PMC2612334. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=58149395090&origin=inward.15. Camilleri S, McDonald F. 2006; Runx2 and dental development. Eur J Oral Sci. 114:361–373. DOI: 10.1111/j.1600-0722.2006.00399.x. PMID: 17026500. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=33749516799&origin=inward.16. Xu Y, Zhao W, Olson SD, Prabhakara KS, Zhou X. 2018; Alternative splicing links histone modifications to stem cell fate decision. Genome Biol. 19:133. DOI: 10.1186/s13059-018-1512-3. PMID: 30217220. PMCID: PMC6138936. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85053251288&origin=inward.17. Agosto LM, Lynch KW. 2018; Alternative pre-mRNA splicing switch controls hESC pluripotency and differentiation. Genes Dev. 32:1103–1104. DOI: 10.1101/gad.318451.118. PMID: 30181358. PMCID: PMC6120719. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85053278380&origin=inward.18. Liu L, Huang R, Yang R, Wei X. 2017; OCT4B1 regulates the cellular stress response of human dental pulp cells with inflammation. Biomed Res Int. 2017:2756891. DOI: 10.1155/2017/2756891. PMID: 28473980. PMCID: PMC5394356. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85018858693&origin=inward.19. Busch A, Hertel KJ. 2012; Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev RNA. 3:1–12. DOI: 10.1002/wrna.100. PMID: 21898828. PMCID: PMC3235224. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84863255704&origin=inward.20. Jeong S. 2017; SR proteins: binders, regulators, and connectors of RNA. Mol Cells. 40:1–9. DOI: 10.14348/molcells.2017.2319. PMID: 28152302. PMCID: PMC5303883. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85018478596&origin=inward.21. Wei WJ, Mu SR, Heiner M, Fu X, Cao LJ, Gong XF, Bindereif A, Hui J. 2012; YB-1 binds to CAUC motifs and stimulates exon inclusion by enhancing the recruitment of U2AF to weak polypyrimidine tracts. Nucleic Acids Res. 40:8622–8636. DOI: 10.1093/nar/gks579. PMID: 22730292. PMCID: PMC3458536. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84866927694&origin=inward.22. Suresh PS, Tsutsumi R, Venkatesh T. 2018; YBX1 at the crossroads of non-coding transcriptome, exosomal, and cytoplasmic granular signaling. Eur J Cell Biol. 97:163–167. DOI: 10.1016/j.ejcb.2018.02.003. PMID: 29478751. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85042379594&origin=inward.23. Mordovkina D, Lyabin DN, Smolin EA, Sogorina EM, Ovchinnikov LP, Eliseeva I. 2020; Y-box binding proteins in mRNP assembly, translation, and stability control. Biomo-lecules. 10:591. DOI: 10.3390/biom10040591. PMID: 32290447. PMCID: PMC7226217. PMID: 307e504e3b15478e8655c5518aaabfa8. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85083404409&origin=inward.24. Lin H, Xu L, Liu H, Sun Q, Chen Z, Yuan G, Chen Z. 2011; KLF4 promotes the odontoblastic differentiation of human dental pulp cells. J Endod. 37:948–954. DOI: 10.1016/j.joen.2011.03.030. PMID: 21689550. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=79959374884&origin=inward.25. Shi JH, Zheng B, Chen S, Ma GY, Wen JK. 2012; Retinoic acid receptor α mediates all-trans-retinoic acid-induced Klf4 gene expression by regulating Klf4 promoter activity in vascular smooth muscle cells. J Biol Chem. 287:10799–10811. DOI: 10.1074/jbc.M111.321836. PMID: 22337869. PMCID: PMC3322846. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84859488310&origin=inward.26. Alkrekshi A, Wang W, Rana PS, Markovic V, Sossey-Alaoui K. 2021; A comprehensive review of the functions of YB-1 in cancer stemness, metastasis and drug resistance. Cell Signal. 85:110073. DOI: 10.1016/j.cellsig.2021.110073. PMID: 34224843. PMCID: PMC8878385. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85110136124&origin=inward.27. Pratap J, Galindo M, Zaidi SK, Vradii D, Bhat BM, Robinson JA, Choi JY, Komori T, Stein JL, Lian JB, Stein GS, van Wijnen AJ. 2003; Cell growth regulatory role of Runx2 during proliferative expansion of preosteoblasts. Cancer Res. 63:5357–5362. PMID: 14500368. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0141705433&origin=inward.28. Wysokinski D, Blasiak J, Pawlowska E. 2015; Role of RUNX2 in breast carcinogenesis. Int J Mol Sci. 16:20969–20993. DOI: 10.3390/ijms160920969. PMID: 26404249. PMCID: PMC4613236. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84940863220&origin=inward.29. Ozaki T, Wu D, Sugimoto H, Nagase H, Nakagawara A. 2013; Runt-related transcription factor 2 (RUNX2) inhibits p53-dependent apoptosis through the collaboration with HDAC6 in response to DNA damage. Cell Death Dis. 4:e610. DOI: 10.1038/cddis.2013.127. PMID: 23618908. PMCID: PMC3641350. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84876924646&origin=inward.30. Schittek B, Psenner K, Sauer B, Meier F, Iftner T, Garbe C. 2007; The increased expression of Y box-binding protein 1 in melanoma stimulates proliferation and tumor invasion, antagonizes apoptosis and enhances chemoresistance. Int J Cancer. 120:2110–2118. DOI: 10.1002/ijc.22512. PMID: 17266041. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=34147097875&origin=inward.31. Homer C, Knight DA, Hananeia L, Sheard P, Risk J, Lasham A, Royds JA, Braithwaite AW. 2005; Y-box factor YB1 controls p53 apoptotic function. Oncogene. 24:8314–8325. DOI: 10.1038/sj.onc.1208998. PMID: 16158057. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=32844472704&origin=inward.32. Oren A, Toporik A, Biton S, Almogy N, Eshel D, Bernstein J, Savitsky K, Rotman G. 2004; hCHL2, a novel chordin-related gene, displays differential expression and complex alternative splicing in human tissues and during myoblast and osteoblast maturation. Gene. 331:17–31. DOI: 10.1016/j.gene.2004.01.029. PMID: 15094188. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=16544394177&origin=inward.33. Jiang Q, Qin X, Kawane T, Komori H, Matsuo Y, Taniuchi I, Ito K, Izumi S, Komori T. 2016; Cbfb2 isoform dominates more potent Cbfb1 and is required for skeletal development. J Bone Miner Res. 31:1391–1404. DOI: 10.1002/jbmr.2814. PMID: 26890219. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84977578612&origin=inward.34. Kazantseva J, Kivil A, Tints K, Kazantseva A, Neuman T, Palm K. 2013; Alternative splicing targeting the hTAF4-TAFH domain of TAF4 represses proliferation and accelerates chondrogenic differentiation of human mesenchymal stem cells. PLoS One. 8:e74799. DOI: 10.1371/journal.pone.0074799. PMID: 24098348. PMCID: PMC3788782. PMID: 471ee20f209a4e77b92458387e0802df. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84884886677&origin=inward.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- MiR-148a-3p Regulates the Invasion and Odontoblastic Differentiation of Human Dental Pulp Stem Cells via the Wnt1/β-Catenin Pathway
- Comparison of Gene Expression from Supernumerary Dental Pulp and Periodontal Ligament Stem Cells
- Dental Pulp Stem Cells and Current in vivo Approaches to Study Dental Pulp Stem Cells in Pulp Injury and Regeneration
- Advances in Research on Stem Cell-Based Pulp Regeneration
- Characterization of Human Dental Pulp Cells from Supernumerary Teeth by Using Flow Cytometry Analysis