Cancer Res Treat.
2022 Jul;54(3):690-708. 10.4143/crt.2021.1121.
Hypo-trimethylation of Histone H3 Lysine 4 and Hyper-tri/dimethylation of Histone H3 Lysine 27 as Epigenetic Markers of Poor Prognosis in Patients with Primary Central Nervous System Lymphoma
- Affiliations
-
- 1Division of Neuro-Oncology and Department of Neurosurgery, Samsung Changwon Hospital, Sungkyunkwan University School of Medicine, Changwon, Korea
- 2Department of New Biology, Daegu Gyeongbuk Institute of Science and Technology, Daegu, Korea
- 3Translational Responsive Medicine Center, Daegu Gyeongbuk Institute of Science and Technology, Daegu, Korea
- 4Well Aging Research Center, Division of Biotechnology, Daegu Gyeongbuk Institute of Science and Technology, Daegu, Korea
- 5Department of Pathology, Samsung Changwon Hospital, Sungkyunkwan University School of Medicine, Changwon, Korea
- 6Department of Pathology, Dong-A University Hospital, Dong-A University College of Medicine, Busan, Korea
- 7Cancer Research Institute, Clinomics Inc., Suwon, Korea
- KMID: 2531316
- DOI: http://doi.org/10.4143/crt.2021.1121
Abstract
- Purpose
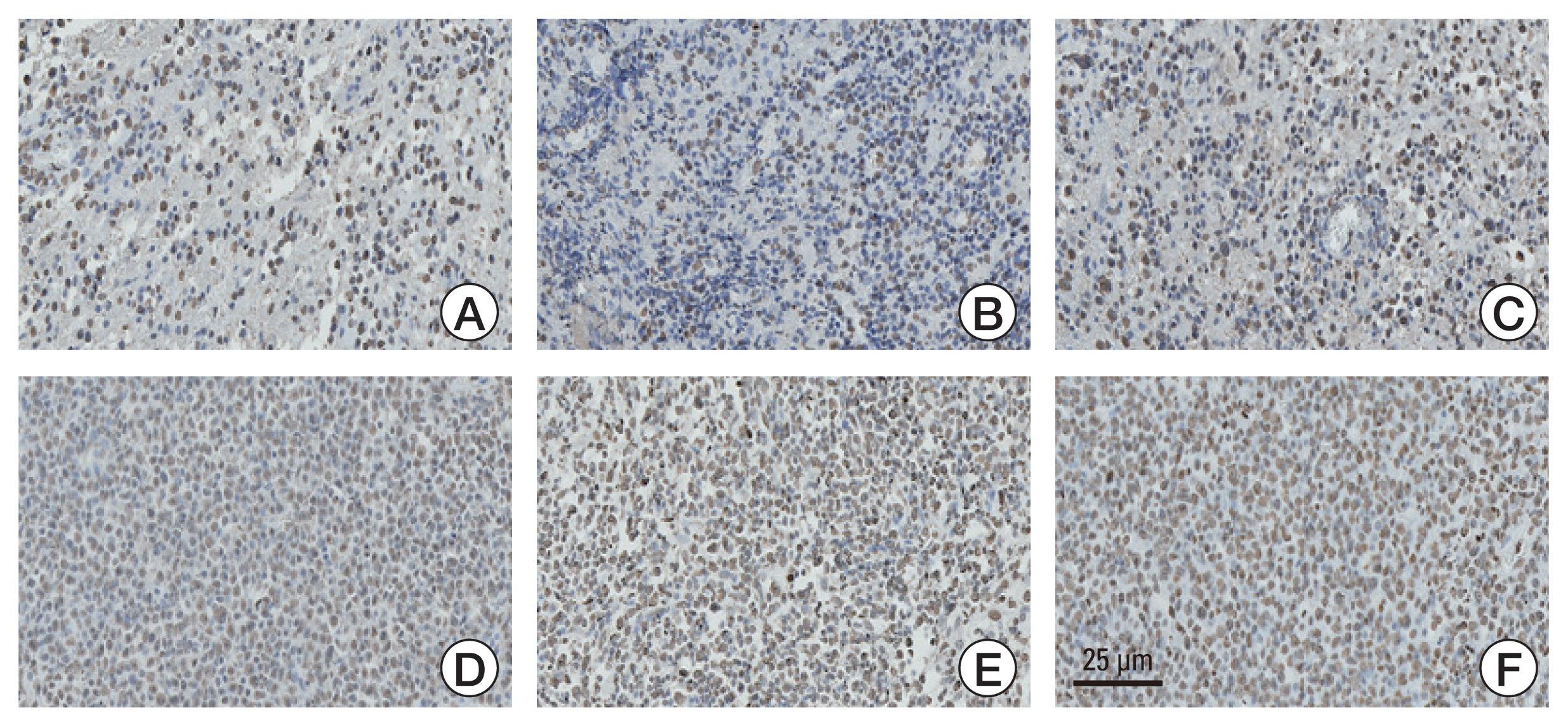
This study aimed to investigate the methylation status of major histone modification sites in primary central nervous system lymphoma (PCNSL) samples and examine their prognostic roles in patients with PCNSL. Materials and Method Between 2007 and 2020, 87 patients were histopathologically diagnosed with PCNSL. We performed immunohistochemical staining of the formalin-fixed paraffin-embedded samples of PCNSL for major histone modification sites, such as H3K4, H3K9, H3K27, H3K14, and H3K36. After detection of meaningful methylation sites, we examined histone modification enzymes that induce methylation or demethylation at each site using immunohistochemical staining. The meaningful immunoreactivity was validated by western blotting using fresh tissue of PCNSL.
Results
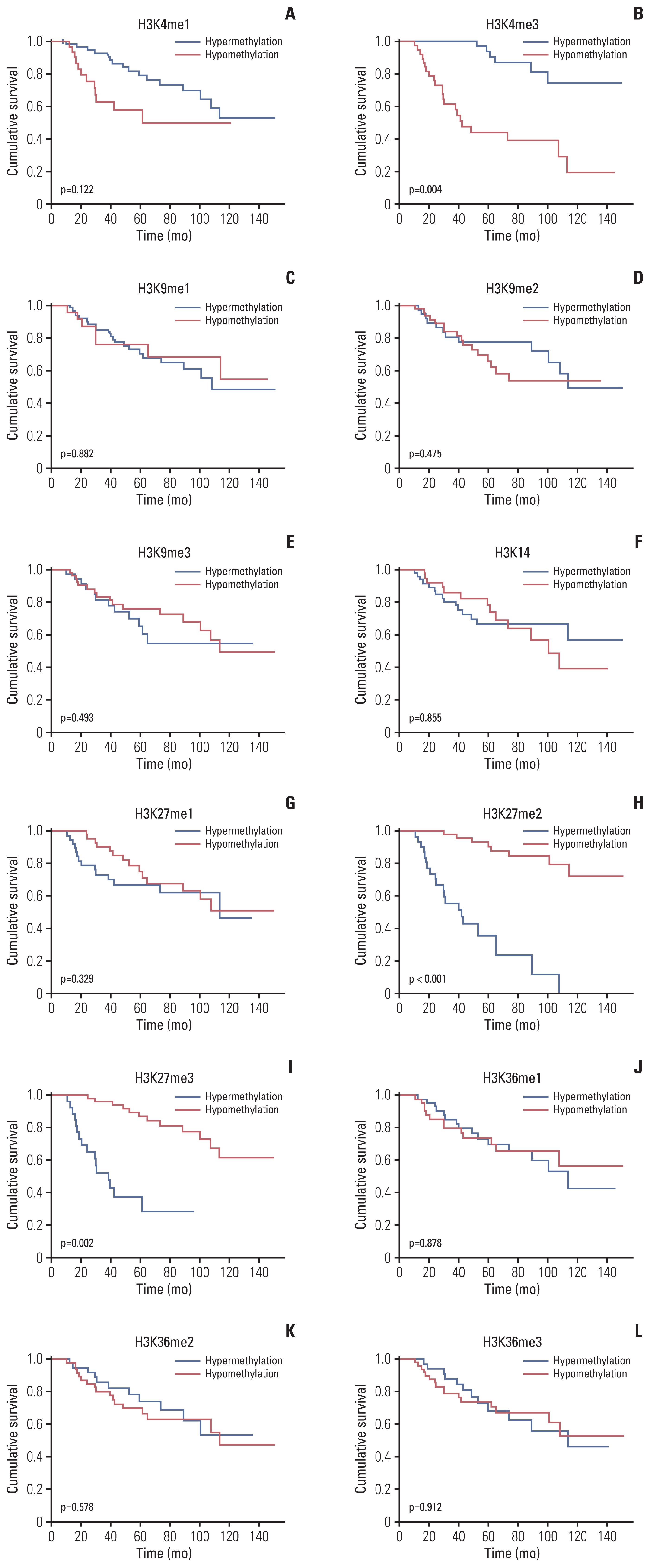
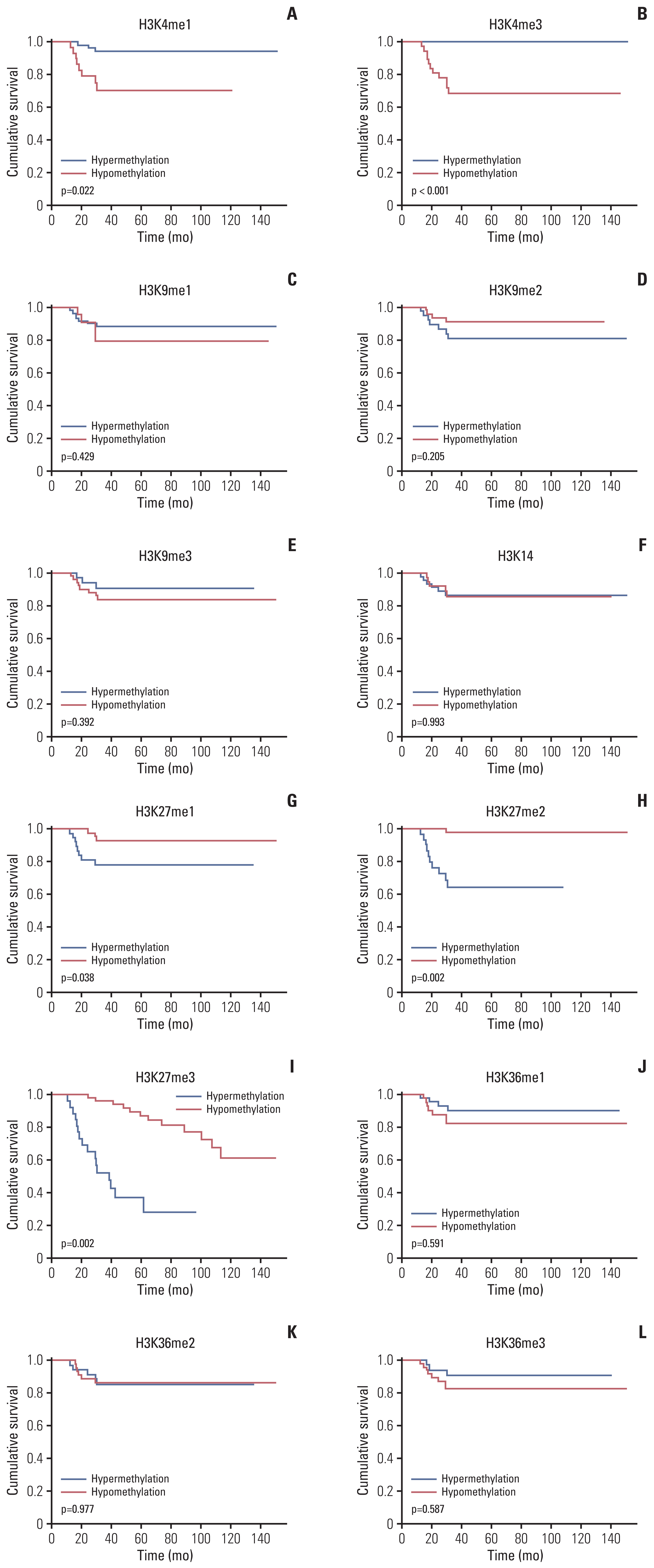
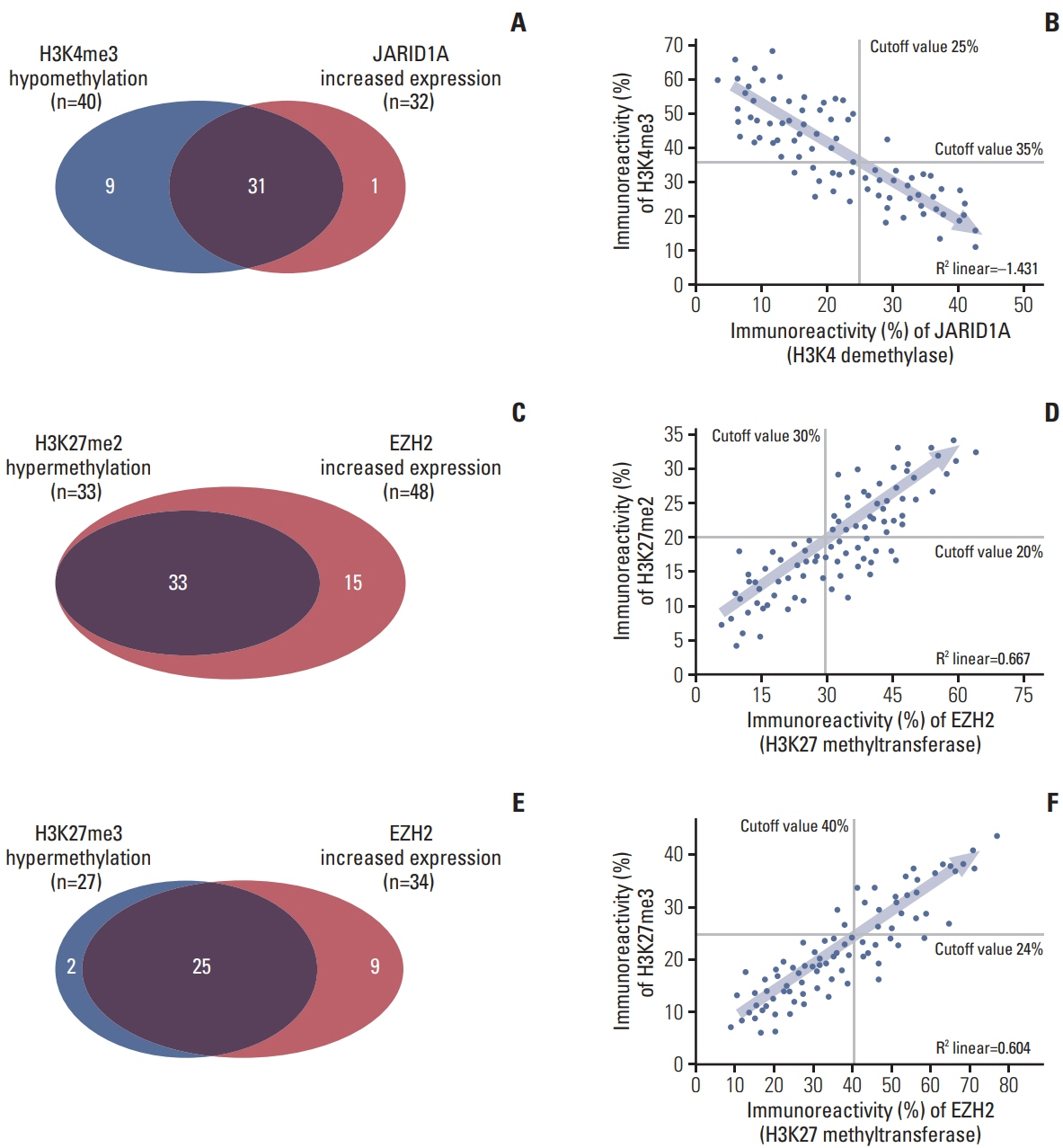
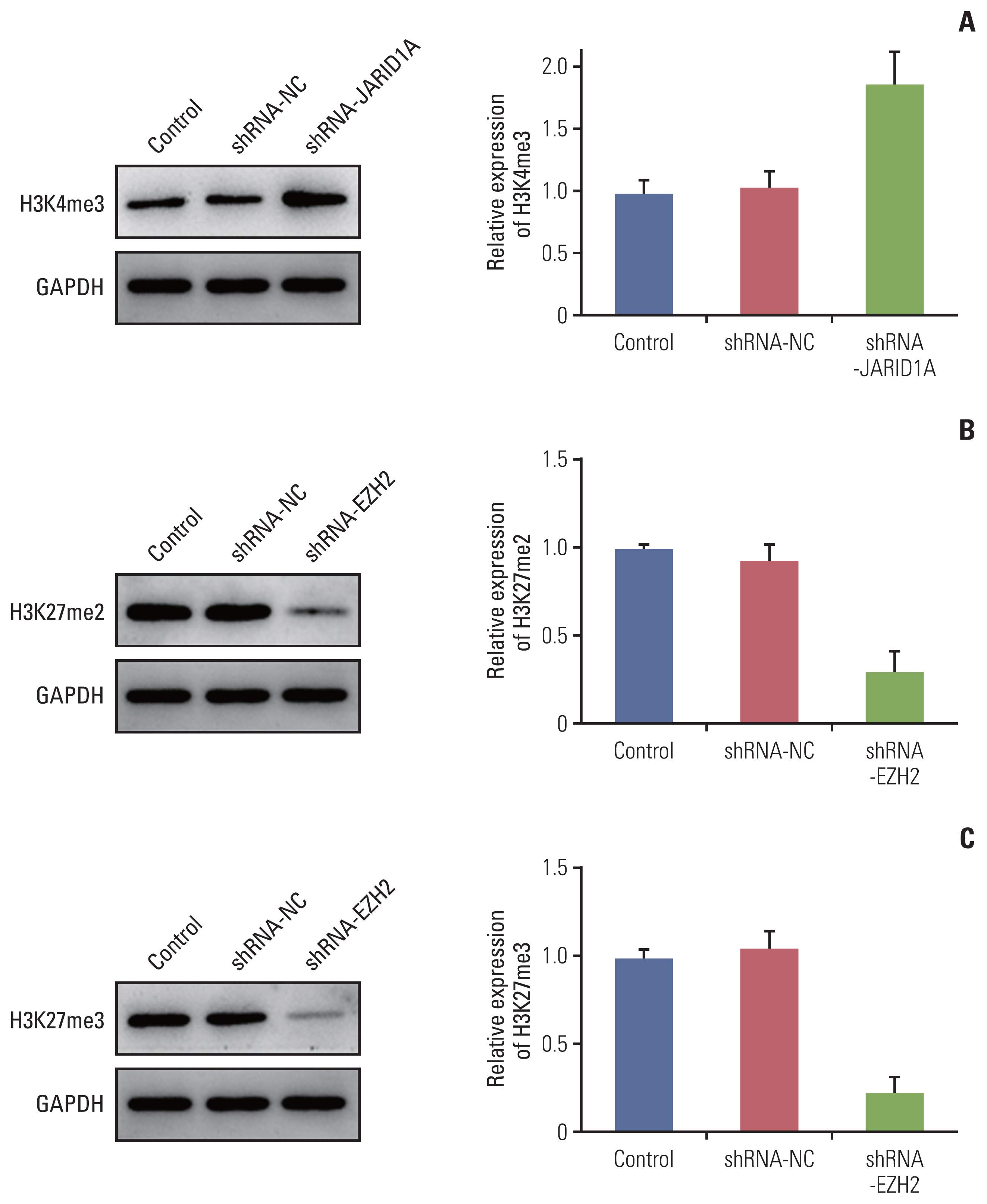
More frequent recurrences were found in hypomethylation of H3K4me3 (p=0.004) and hypermethylation of H3K27me2 (p<0.001) and H3K27me3 (p=0.002). These factors were also statistically related to short PFS and overall survival in the univariate and multivariate analyses. Next, histone modification enzymes inducing the demethylation of H3K4 (lysine-specific demethylase-1/2 and Jumonji AT-rich interactive domain [JARID] 1A-D]) and methylation of H3K27 (enhancer of zeste homolog [EZH]-1/2) were immu- nohistochemically stained. Among them, the immunoreactivity of JARID1A inversely associated with the methylation status of H3K4me3 (R2=-1.431), and immunoreactivity of EZH2 was directly associated with the methylation status of H3K27me2 (R2=0.667) and H3K27me3 (R2=0.604). These results were validated by western blotting in fresh PCNSL samples.
Conclusion
Our study suggests that hypomethylation of H3K4me3 and hypermethylation of H3K27me2 and H3K27me3 could be associated with poor outcomes in patients with PCNSL and that these relationships are modified by JARID1A and EZH2.
Keyword
Figure
Reference
-
References
1. Dho YS, Jung KW, Ha J, Seo Y, Park CK, Won YJ, et al. An updated nationwide epidemiology of primary brain tumors in Republic of Korea, 2013. Brain Tumor Res Treat. 2017; 5:16–23.
Article2. O’Neill BP, Decker PA, Tieu C, Cerhan JR. The changing incidence of primary central nervous system lymphoma is driven primarily by the changing incidence in young and middle-aged men and differs from time trends in systemic diffuse large B-cell non-Hodgkin’s lymphoma. Am J Hematol. 2013; 88:997–1000.
Article3. Mead GM, Bleehen NM, Gregor A, Bullimore J, Shirley D, Rampling RP, et al. A medical research council randomized trial in patients with primary cerebral non-Hodgkin lymphoma: cerebral radiotherapy with and without cyclophosphamide, doxorubicin, vincristine, and prednisone chemotherapy. Cancer. 2000; 89:1359–70.
Article4. Hoang-Xuan K, Bessell E, Bromberg J, Hottinger AF, Preusser M, Ruda R, et al. Diagnosis and treatment of primary CNS lymphoma in immunocompetent patients: guidelines from the European Association for Neuro-Oncology. Lancet Oncol. 2015; 16:e322–32.
Article5. Ferreri AJ, Blay JY, Reni M, Pasini F, Spina M, Ambrosetti A, et al. Prognostic scoring system for primary CNS lymphomas: the International Extranodal Lymphoma Study Group experience. J Clin Oncol. 2003; 21:266–72.
Article6. King RL, Goodlad JR, Calaminici M, Dotlic S, Montes-Moreno S, Oschlies I, et al. Lymphomas arising in immune-privileged sites: insights into biology, diagnosis, and pathogenesis. Virchows Arch. 2020; 476:647–65.
Article7. Bakhshi TJ, Georgel PT. Genetic and epigenetic determinants of diffuse large B-cell lymphoma. Blood Cancer J. 2020; 10:123.
Article8. Lue JK, Amengual JE. Emerging EZH2 inhibitors and their application in lymphoma. Curr Hematol Malig Rep. 2018; 13:369–82.
Article9. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016; 127:2391–405.
Article10. Graham MS, Mellinghoff IK. Histone-mutant glioma: molecular mechanisms, preclinical models, and implications for therapy. Int J Mol Sci. 2020; 21:7193.
Article11. Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012; 482:226–31.12. Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012; 44:251–3.
Article13. Li B, Chng WJ. EZH2 abnormalities in lymphoid malignancies: underlying mechanisms and therapeutic implications. J Hematol Oncol. 2019; 12:118.
Article14. Beguelin W, Rivas MA, Calvo Fernandez MT, Teater M, Purwada A, Redmond D, et al. EZH2 enables germinal centre formation through epigenetic silencing of CDKN1A and an Rb-E2F1 feedback loop. Nat Commun. 2017; 8:877.
Article15. Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. 2018; 378:1396–407.
Article16. Wagener N, Macher-Goeppinger S, Pritsch M, Husing J, Hoppe-Seyler K, Schirmacher P, et al. Enhancer of zeste homolog 2 (EZH2) expression is an independent prognostic factor in renal cell carcinoma. BMC Cancer. 2010; 10:524.
Article17. Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011; 476:298–303.
Article18. Zhao X, Lwin T, Zhang X, Huang A, Wang J, Marquez VE, et al. Disruption of the MYC-miRNA-EZH2 loop to suppress aggressive B-cell lymphoma survival and clonogenicity. Leukemia. 2013; 27:2341–50.
Article19. Tateishi K, Miyake Y, Nakamura T, Yamamoto T. Primary central nervous system lymphoma: clinicopathological and genomic insights for therapeutic development. Brain Tumor Pathol. 2021; 38:173–82.
Article20. Murata T, Kondo Y, Sugimoto A, Kawashima D, Saito S, Isomura H, et al. Epigenetic histone modification of Epstein-Barr virus BZLF1 promoter during latency and reactivation in Raji cells. J Virol. 2012; 86:4752–61.
Article21. de Oliveira DE, Ballon G, Cesarman E. NF-kappaB signaling modulation by EBV and KSHV. Trends Microbiol. 2010; 18:248–57.22. Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, et al. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007; 128:1063–76.
Article23. Mao S, Neale GA, Goorha RM. T-cell oncogene rhombotin-2 interacts with retinoblastoma-binding protein 2. Oncogene. 1997; 14:1531–9.
Article24. Yang GJ, Zhu MH, Lu XJ, Liu YJ, Lu JF, Leung CH, et al. The emerging role of KDM5A in human cancer. J Hematol Oncol. 2021; 14:30.
Article25. Harmeyer KM, Facompre ND, Herlyn M, Basu D. JARID1 histone demethylases: emerging targets in cancer. Trends Cancer. 2017; 3:713–25.
Article26. Hou J, Wu J, Dombkowski A, Zhang K, Holowatyj A, Boerner JL, et al. Genomic amplification and a role in drug-resistance for the KDM5A histone demethylase in breast cancer. Am J Transl Res. 2012; 4:247–56.27. Easwaran H, Tsai HC, Baylin SB. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol Cell. 2014; 54:716–27.
Article28. Hyun K, Jeon J, Park K, Kim J. Writing, erasing and reading histone lysine methylations. Exp Mol Med. 2017; 49:e324.
Article29. Sheikhzadeh F, Ward RK, van Niekerk D, Guillaud M. Automatic labeling of molecular biomarkers of immunohistochemistry images using fully convolutional networks. PLoS One. 2018; 13:e0190783.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Global Histone Modification Patterns of Osteosarcoma
- Writing, erasing and reading histone lysine methylations
- Inhibition of Nuclear Receptor Binding SET Domain 2/Multiple Myeloma SET Domain by LEM-06 Implication for Epigenetic Cancer Therapies
- Histone lysine demethylases in mammalian embryonic development
- Epigenetic Regulation of Nuclear Factor Erythroid-2-Related Factor 2 in Colorectal Cancer Cells Resistant to Ionizing Radiation