Healthc Inform Res.
2022 Jan;28(1):16-24. 10.4258/hir.2022.28.1.16.
Protected Health Information Recognition by Fine-Tuning a Pre-training Transformer Model
- Affiliations
-
- 1Department of IT Convergence Engineering, Gachon University, Seongnam, Korea
- 2Department of Computer Engineering, Gachon University, Seongnam, Korea
- KMID: 2526490
- DOI: http://doi.org/10.4258/hir.2022.28.1.16
Abstract
Objectives
De-identifying protected health information (PHI) in medical documents is important, and a prerequisite to deidentification is the identification of PHI entity names in clinical documents. This study aimed to compare the performance of three pre-training models that have recently attracted significant attention and to determine which model is more suitable for PHI recognition.
Methods
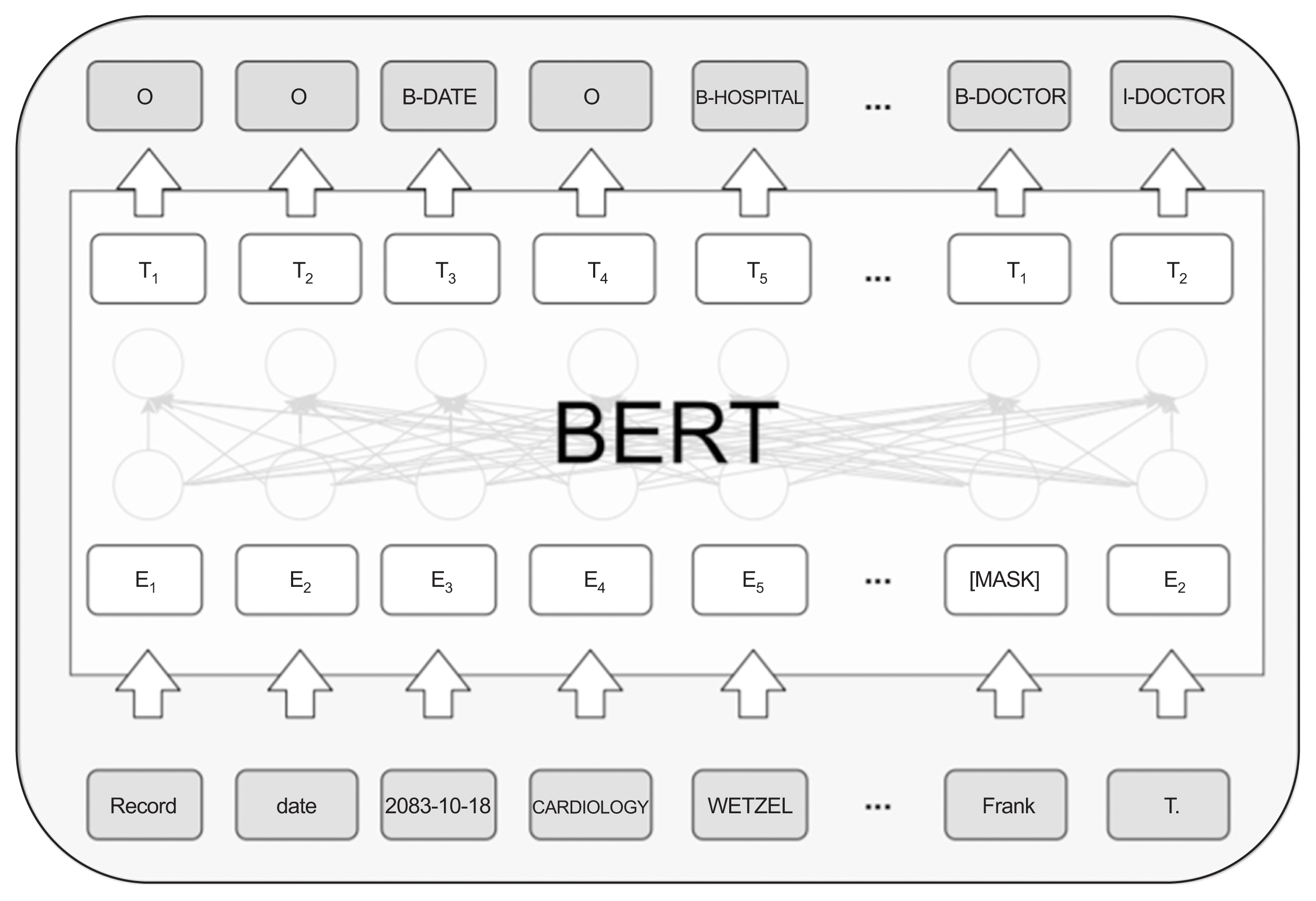
We compared the PHI recognition performance of deep learning models using the i2b2 2014 dataset. We used the three pre-training models—namely, bidirectional encoder representations from transformers (BERT), robustly optimized BERT pre-training approach (RoBERTa), and XLNet (model built based on Transformer-XL)—to detect PHI. After the dataset was tokenized, it was processed using an inside-outside-beginning tagging scheme and WordPiecetokenized to place it into these models. Further, the PHI recognition performance was investigated using BERT, RoBERTa, and XLNet.
Results
Comparing the PHI recognition performance of the three models, it was confirmed that XLNet had a superior F1-score of 96.29%. In addition, when checking PHI entity performance evaluation, RoBERTa and XLNet showed a 30% improvement in performance compared to BERT.
Conclusions
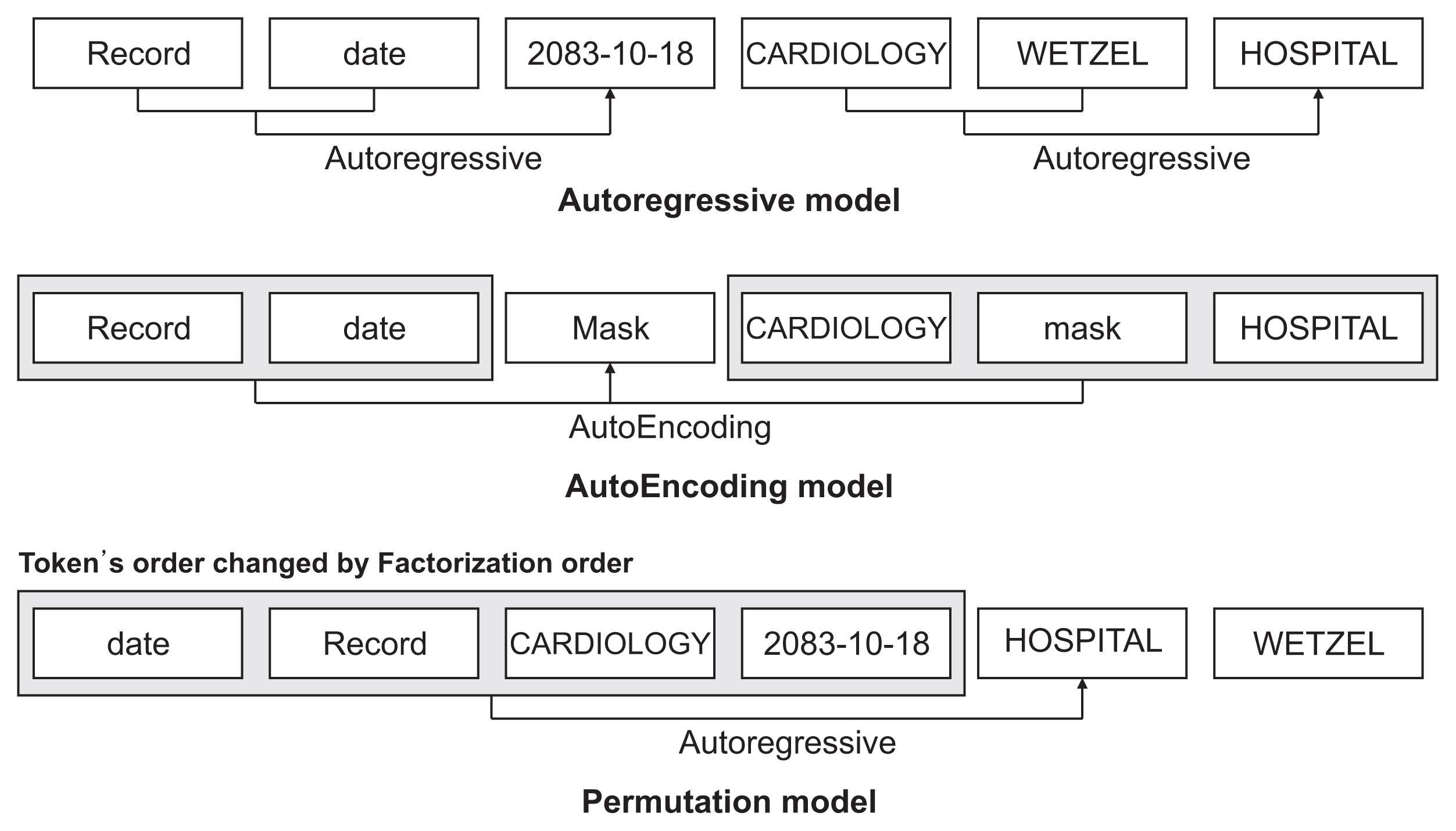
Among the pre-training models used in this study, XLNet exhibited superior performance because word embedding was well constructed using the two-stream self-attention method. In addition, compared to BERT, RoBERTa and XLNet showed superior performance, indicating that they were more effective in grasping the context.
Figure
Reference
-
References
1. Park YT, Kim YS, Yi BK, Kim SM. Clinical decision support functions and digitalization of clinical documents of electronic medical record systems. Healthc Inform Res. 2019; 25(2):115–23.
Article2. Choi YI, Park SJ, Chung JW, Kim KO, Cho JH, et al. Development of machine learning model to predict the 5-year risk of starting biologic agents in patients with inflammatory bowel disease (IBD): K-CDM network study. J Clin Med. 2020; 9(11):3427.
Article3. Seong D, Yi BK. Research trends in clinical natural language processing. Commun Korean Inst Inf Sci Eng. 2017; 35(5):20–6.4. Shin SY. Privacy protection and data utilization. Healthc Inform Res. 2021; 27(1):1–2.
Article5. National Committee on Vital and Health Statistics. Health Information Privacy Beyond HIPAA: A 2018 Environmental Scan of Major Trends and Challenges [Internet]. Hyattsville (MD): National Committee on Vital and Health Statistics;2017. [cited at 2022 Jan 10]. Available from: https://ncvhs.hhs.gov/wp-content/uploads/2018/02/NCVHS-Beyond-HIPAA_Report-Final-02-08-18.pdf .6. Shin SY, Park YR, Shin Y, Choi HJ, Park J, Lyu Y, et al. A de-identification method for bilingual clinical texts of various note types. J Korean Med Sci. 2015; 30(1):7–15.
Article7. Lafferty J, McCallum A, Pereira FC. Conditional random fields: probabilistic models for segmenting and labeling sequence data. In : Proceedings of the 18th International Conference on Machine Learning (ICML); 2001 Jun 28–Jul 1; San Francisco, CA. p. 282–9.8. Wang Y. Annotating and recognising named entities in clinical notes. In : Proceedings of the ACL-IJCNLP 2009 Student Research Workshop; 2009 Aug 4; Suntec, Singapore. p. 18–26.
Article9. Dreyfus SE. Artificial neural networks, back propagation, and the Kelley-Bryson gradient procedure. J Guid Control Dyn. 1990; 13(5):926–8.
Article10. Team AI Korea. Recurrent neural network (RNN) tutorial, Part 1 [Internet]. [place unknow]: Team AI Korea;2015. [cited at 2022 Jan 10]. Available from: http://aikorea.org/blog/rnn-tutorial-1/ .11. Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput. 1997; 9(8):1735–80.
Article12. Liu Z, Yang M, Wang X, Chen Q, Tang B, Wang Z, et al. Entity recognition from clinical texts via recurrent neural network. BMC Med Inform Decis Mak. 2017; 17(Suppl 2):67.
Article13. Mikolov T, Chen K, Corrado G, Dean J. Efficient estimation of word representations in vector space [Internet]. Ithaca (NY): arXiv.org;2013. [cited at 2022 Jan 10]. Available from: https://arxiv.org/abs/1301.3781 .14. fastText [Internet]. Menlo Park (CA): Facebook Inc.;2020. [cited at 2022 Jan 10]. Available from: https://fast-text.cc/ .15. Peters ME, Neumann M, Iyyer M, Gardner M, Clark C, Lee K, et al. Deep contextualized word representations [Internet]. Ithaca (NY): arXiv.org;2018. [cited at 2022 Jan 10]. Available from: https://arxiv.org/abs/1802.05365 .16. Kim JM, Lee JH. Text document classification based on recurrent neural network using word2vec. J Korean Inst Intell Syst. 2017; 27(6):560–5.
Article17. Stubbs A, Kotfila C, Uzuner O. Automated systems for the de-identification of longitudinal clinical narratives: overview of 2014i2b2/UTHealth shared task Track 1. J Biomed Inform. 2015; 58 Suppl. (Suppl):S11–S19.18. DBMI Data Portal. n2c2 NLP Research Data Sets [Internet]. Boston (MA): Harvard Medical School;2019. [cited at 2022 Jan 10]. Available from: https://portal.dbmi.hms.harvard.edu/projects/n2c2-nlp/ .19. Devlin J, Chang MW, Lee K, Toutanova K. BERT: pre-training of deep bidirectional transformers for language understanding [Internet]. Ithaca (NY): arXiv.org;2018. [cited at 2022 Jan 10]. Available from: https://arxiv.org/abs/1810.04805 .
Article20. Lan Z, Chen M, Goodman S, Gimpel K, Sharma P, Soricut R. ALBERT: a lite BERT for self-supervised learning of language representations. In : Proceedings of the 8th International Conference on Learning Representations (ICLR); 2020 Apr 26–30; Addis Ababa, Ethiopia.21. Liu Y, Ott M, Goyal N, Du J, Joshi M, Chen D, et al. ROBERTa: a robustly optimized BERT pretraining approach [Internet]. Ithaca (NY): arXiv.org;2019. [cited at 2022 Jan 10]. Available from: https://arxiv.org/abs/1907.11692 .22. Hugging Face. deepset/roberta-base-squad2 [Internet]. New York (NY): Hugging Face;2020. [cited at 2022 Jan 10]. Available from: https://huggingface.co/deepset/roberta-base-squad2 .23. Kingma DP, Ba J. Adam: a method for stochastic optimization [Internet]. Ithaca (NY): arXiv.org;2014. [cited at 2022 Jan 10]. Available from: https://arxiv.org/abs/1412.6980 .24. Yang Z, Dai Z, Yang Y, Carbonell J, Salakhutdinov RR, Le QV. Xlnet: generalized autoregressive pretraining for language understanding. Adv Neural Inf Process Syst. 2019; 32:5754–64.25. Hugging Face. xlnet-base-cased [Internet]. New York (NY): Hugging Face;2019. [cited at 2022 Jan 10]. Available from: https://huggingface.co/xlnet-base-cased .26. Warby SC, Wendt SL, Welinder P, Munk EG, Carrillo O, Sorensen HB, et al. Sleep-spindle detection: crowdsourcing and evaluating performance of experts, non-experts and automated methods. Nat Methods. 2014; 11(4):385–92.
Article27. Kim YW, Cho N, Jang HJ. Trends in Research on the security of medical information in Korea: focused on information privacy security in hospitals. Healthc Inform Res. 2018; 24(1):61–8.
Article28. Kingma DP, Welling M. Auto-encoding variational bayes [Internet]. Ithaca (NY): arXiv.org;2013. [cited at 2022 Jan 10]. Available from: https://arxiv.org/abs/1312.6114 .29. Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial nets. Adv Neural Inf Process Syst. 2014; 27:2672–80.30. Alsentzer E, Murphy JR, Boag W, Weng WH, Jin D, Naumann T, et al. Publicly available clinical BERT embeddings [Internet]. Ithaca (NY): arXiv.org;2019. [cited at 2022 Jan 10]. Available from: https://arxiv.org/abs/1904.03323 .
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Large Language Models: A Guide for Radiologists
- Deep Learning-Enabled Detection of Pneumoperitoneum in Supine and Erect Abdominal Radiography: Modeling Using Transfer Learning and Semi-Supervised Learning
- Utilizing ChatGPT in clinical research related to anesthesiology: a comprehensive review of opportunities and limitations
- Strategies for Protecting the Privacy in Genetic Testing
- Can We Trust the Information on Allergic Rhinitis From Chat Generated Pre-Trained Transformer?