Ann Surg Treat Res.
2021 Sep;101(3):140-150. 10.4174/astr.2021.101.3.140.
Circulating microRNAs as biomarkers in bile-derived exosomes of cholangiocarcinoma
- Affiliations
-
- 1Division of Hepatobiliary and Pancreatic Surgery, Department of Surgery, Keimyung University Dongsan Medical Center, Daegu, Korea
- 2Department of Microbiology, Keimyung University School of Medicine, Daegu, Korea
- KMID: 2519844
- DOI: http://doi.org/10.4174/astr.2021.101.3.140
Abstract
- Purpose
In this pilot study, using next-generation sequencing and integrated messenger RNA (mRNA) sequencing, we investigated circulating microRNA (miRNA) expression profiling from bile-derived exosomes to identify dysregulated miRNA signatures and oncogenic pathways and determine their effects on targeted mRNAs in cholangiocarcinoma (CCA). Moreover, we explored the possibility that genetic analysis using bile-derived exosomes may replace gene analysis using tissue.
Methods
Bile was collected from a patient with perihilar CCA before curative resection. As a control, bile was collected from a patient with a common bile duct stone. Exosomes were isolated from the bile, and we performed next-generation miRNA sequencing using isolated exosomes. To evaluate miRNA-mRNA interactions, mRNA sequencing was performed using bile fluid in both patients.
Results
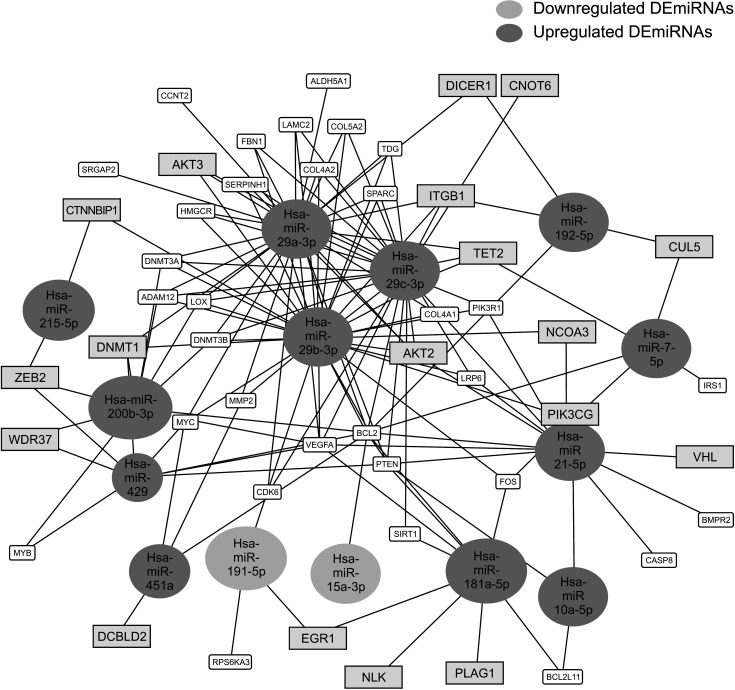
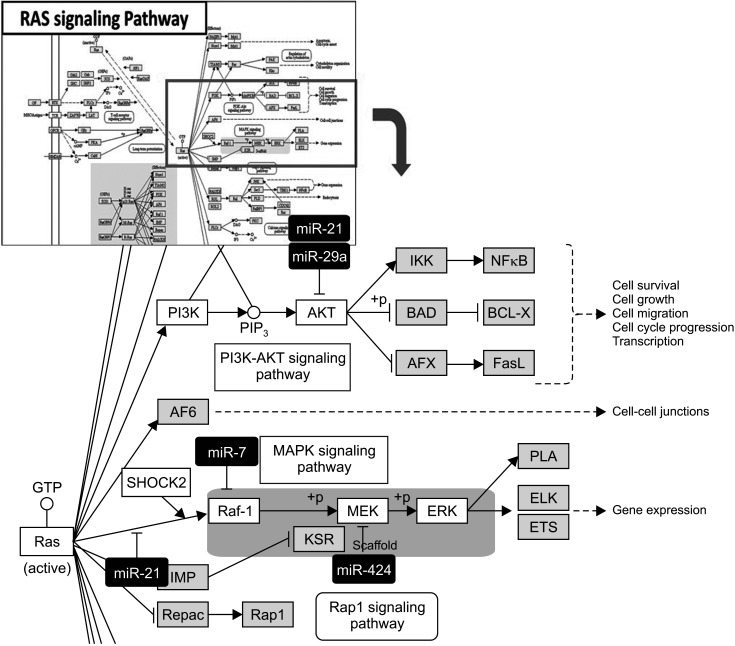
We identified 22 differentially expressed miRNAs. More than 65% of the predicted mRNA targets of those miRNAs were actually differentially expressed between control and CCA bile samples. In functional pathway analysis, targets of 22 miRNAs were primarily enriched in mitogen-activated protein kinase, platelet derived growth factor, vascular endothelial growth factor, epidermal growth factor receptor, and p53 signaling. In particular, in the functional assessment of miRNAmRNA interactions, RAS pathways, including downstream pathways (PI3K-AKT-mTOR and RAS-RAF-MEK-ERK), were determined to be enriched.
Conclusion
Circulating miRNAs in bile-derived exosomes provide new information for the development of miRNA analysis in CCA. These miRNAs may represent the oncogenic characteristics of CCA tissue, enabling them to be used instead of tissue samples for the diagnosis of CCA. Further research investigating circulating miRNAs in bile exosomes may lead to more rational, targeted approaches to treatment.
Keyword
Figure
Reference
-
1. DeOliveira ML, Cunningham SC, Cameron JL, Kamangar F, Winter JM, Lillemoe KD, et al. Cholangiocarcinoma: thirty-one-year experience with 564 patients at a single institution. Ann Surg. 2007; 245:755–762. PMID: 17457168.2. Keller S, Ridinger J, Rupp AK, Janssen JW, Altevogt P. Body f luid derived exosomes as a novel template for clinical diagnostics. J Transl Med. 2011; 9:86. PMID: 21651777.
Article3. Sagredo AI, Sepulveda SA, Roa JC, Oróstica LJ. Exosomes in bile as potential pancreatobiliary tumor biomarkers. Trans Cancer Res. 2017; 6 Suppl 8:S1371–S1383.
Article4. Gobbo J, Marcion G, Cordonnier M, Dias AM, Pernet N, Hammann A, et al. Restoring anticancer immune response by targeting tumor-derived exosomes with a HSP70 peptide aptamer. J Natl Cancer Inst. 2015; 108.
Article5. Subra C, Grand D, Laulagnier K, Stella A, Lambeau G, Paillasse M, et al. Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prost ag landins. J Lipid Res. 2010; 51:2105–2120. PMID: 20424270.6. Prieto D, Sotelo N, Seija N, Sernbo S, Abreu C, Durán R, et al. S100-A9 protein in exosomes from chronic lymphocytic leukemia cells promotes NF-κB activity during disease progression. Blood. 2017; 130:777–788. PMID: 28596424.
Article7. Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019; 38:2844–2859. PMID: 30546088.
Article8. Février B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol. 2004; 16:415–421. PMID: 15261674.
Article9. Kitdumrongthum S, Metheetrairut C, Charoensawan V, Ounjai P, Janpipatkul K, Panvongsa W, et al. Dysregulated microRNA expression prof i les in cholangiocarcinoma cel l -der ived exosomes. Life Sci. 2018; 210:65–75. PMID: 30165035.10. Kosaka N, Yoshioka Y, Fujita Y, Ochiya T. Versatile roles of extracellular vesicles in cancer. J Clin Invest. 2016; 126:1163–1172. PMID: 26974161.
Article11. Li L, Masica D, Ishida M, Tomuleasa C, Umegaki S, Kalloo AN, et al. Human bile contains microRNA-laden extracellular vesicles that can be used for cholangiocarcinoma diagnosis. Hepatology. 2014; 60:896–907. PMID: 24497320.
Article12. Kern F, Fehlmann T, Solomon J, Schwed L, Grammes N, Backes C, et al. miEAA 2.0: integrating multi-species microRNA enrichment analysis and workf low management systems. Nucleic Acids Res. 2020; 48:W521–W528. PMID: 32374865.13. Huangda W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009; 37:1–13. PMID: 19033363.14. Iwakawa HO, Tomari Y. The functions of mic roRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015; 25:651–665. PMID: 26437588.15. Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019; 28:1947–1951. PMID: 31441146.
Article16. Peng Y, Croce CM. The role of microRNAs in human cancer. Signal Transduct Target Ther. 2016; 1:15004. PMID: 29263891.
Article17. Kawahigashi Y, Mishima T, Mizuguchi Y, Arima Y, Yokomuro S, Kanda T, et al. MicroRNA profiling of human intrahepatic cholangiocarcinoma cell lines reveals biliary epithelial cell-specific microRNAs. J Nippon Med Sch. 2009; 76:188–197. PMID: 19755794.
Article18. Plieskatt JL, Rinaldi G, Feng Y, Peng J, Yonglitthipagon P, Easley S, et al. Distinct miRNA signatures associate with subtypes of cholangiocarcinoma from infection with the tumourigenic liver fluke Opisthorchis viverrini. J Hepatol. 2014; 61:850–858. PMID: 25017828.
Article19. Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006; 130:2113–2129. PMID: 16762633.
Article20. Kishimoto T, Eguchi H, Nagano H, Kobayashi S, Akita H, Hama N, et al. Plasma miR-21 is a novel diagnostic biomarker for biliary tract cancer. Cancer Sci. 2013; 104:1626–1631. PMID: 24118467.
Article21. Wang S, Yin J, Li T, Yuan L, Wang D, He J, et al. Upregulated circulating miR-150 is associated with the risk of intrahepatic chol ang ioc arc inoma. Oncol Rep. 2015; 33:819–825. PMID: 25482320.22. Silakit R, Loilome W, Yongvanit P, Chusorn P, Techasen A, Boonmars T, et al. Circulating miR-192 in liver fluke-associated cholangiocarcinoma patients: a prospective prognostic indicator. J Hepatobiliary Pancreat Sci. 2014; 21:864–872. PMID: 25131257.
Article23. Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracel lular communicators in cardiovascular disease. Circ Res. 2012; 110:483–495. PMID: 22302755.24. Zheng B, Jeong S, Zhu Y, Chen L, Xia Q. miRNA and lncRNA as biomarkers in cholangiocarcinoma(CCA). Oncotarget. 2017; 8:100819–100830. PMID: 29246025.
Article25. Selaru FM, Olaru AV, Kan T, David S, Cheng Y, Mori Y, et al. MicroRNA-21 is overexpressed in human cholangiocarcinoma and regulates programmed cell death 4 and tissue inhibitor of metalloproteinase 3. Hepatology. 2009; 49:1595–1601. PMID: 19296468.
Article26. Ahn KS, O'Brien D, Kang YN, Mounajjed T, Kim YH, Kim TS, et al. Prognostic subclass of intrahepatic cholangiocarcinoma by integrative molecular-clinical analysis and potential targeted approach. Hepatol Int. 2019; 13:490–500. PMID: 31214875.
Article27. Andersen JB, Spee B, Blechacz BR, Avital I, Komuta M, Barbour A, et al. Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors. Gastroenterology. 2012; 142:1021–1031. PMID: 22178589.
Article28. Gao L, Yang X, Zhang H, Yu M, Long J, Yang T. Inhibition of miR-10a-5p suppresses cholangiocarcinoma cell growth through downregulation of Akt pathway. Onco Targets Ther. 2018; 11:6981–6994. PMID: 30410355.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Circulating Plasma and Exosomal microRNAs as Indicators of Drug-Induced Organ Injury in Rodent Models
- Exosomes and Microvesicles as Biomarkers in Metabolic Diseases
- Quantitative Analysis of Exosomes From Murine Lung Cancer Cells by Flow Cytometry
- Exosomes as the source of biomarkers of metabolic diseases
- Surveillance and Early Diagnosis of Pancreatic Cancer