J Pathol Transl Med.
2021 Jan;55(1):33-42. 10.4132/jptm.2020.09.29.
Automated immunohistochemical assessment ability to evaluate estrogen and progesterone receptor status compared with quantitative reverse transcription-polymerase chain reaction in breast carcinoma patients
- Affiliations
-
- 1Department of Pathology, Korea University Guro Hospital, Korea University College of Medicine, Seoul, Korea
- KMID: 2511511
- DOI: http://doi.org/10.4132/jptm.2020.09.29
Abstract
- Background
This study aimed to investigate the capability of an automated immunohistochemical (IHC) evaluation of hormonal receptor status in breast cancer patients compared to a well-validated quantitative reverse transcription–polymerase chain reaction (RT-qPCR) method.
Methods
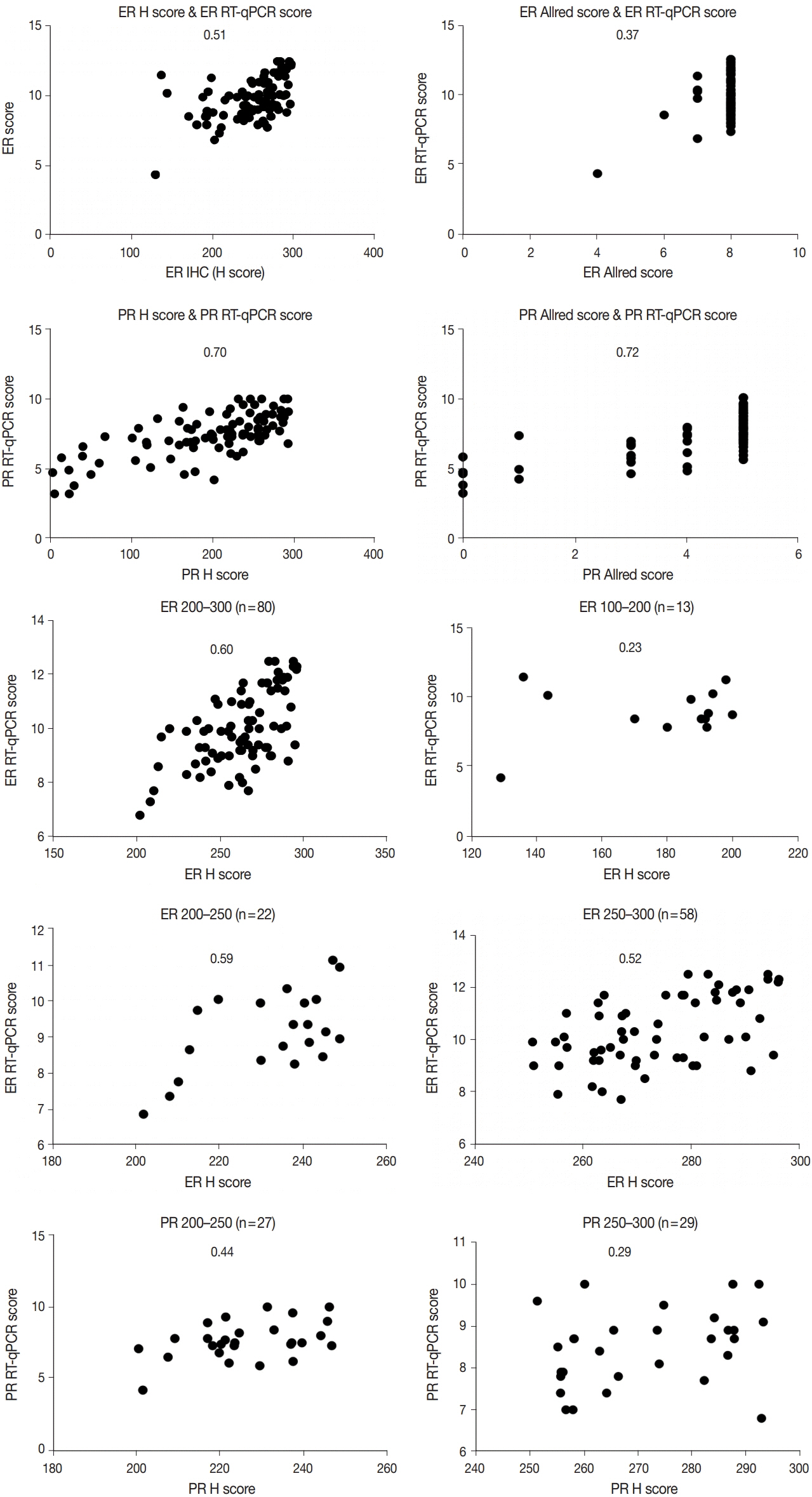
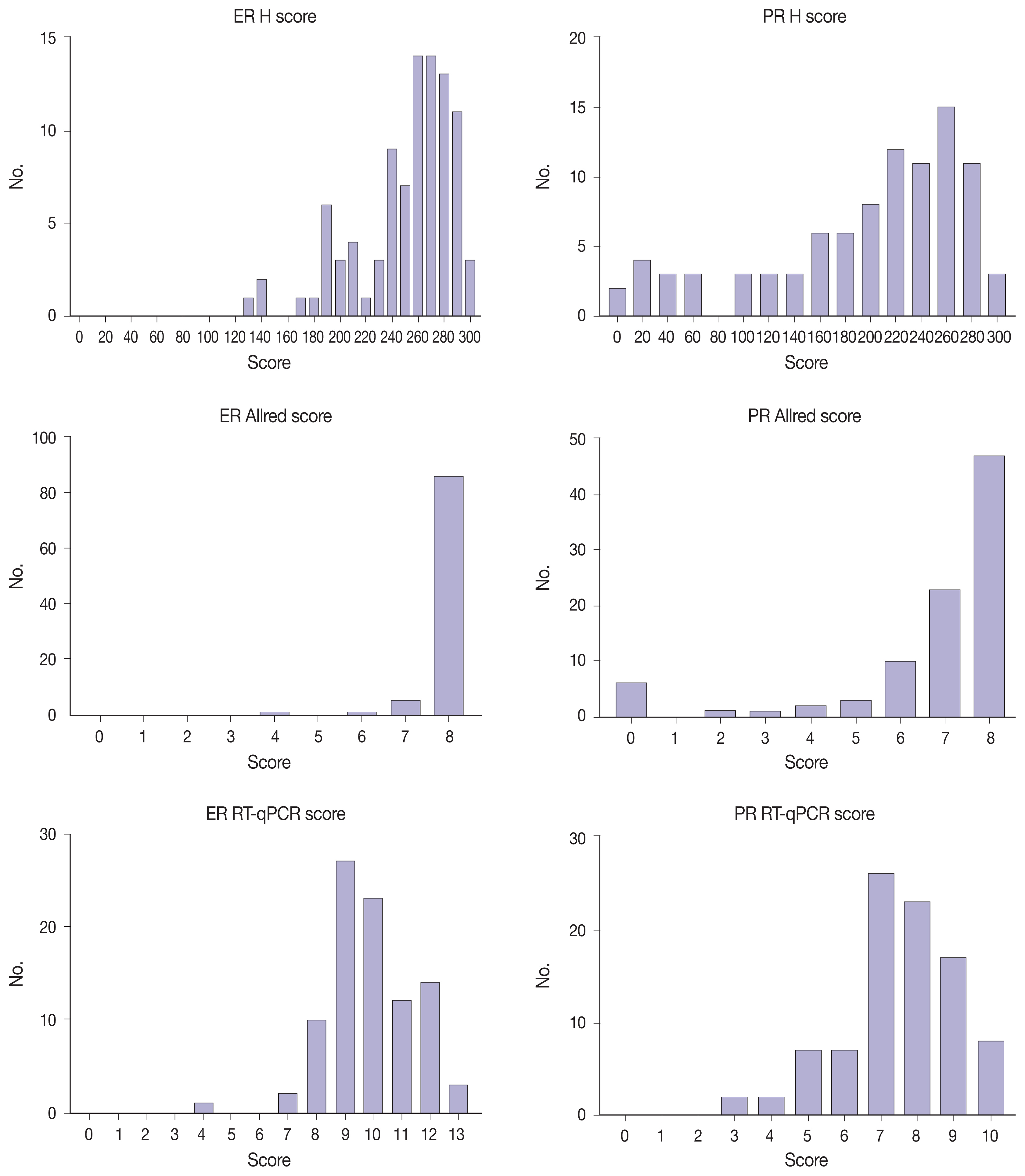
This study included 93 invasive breast carcinoma cases that had both standard IHC assay and Oncotype Dx assay results. The same paraffin blocks on which Oncotype Dx assay had been performed were selected. Estrogen receptor (ER) and progesterone receptor (PR) receptor status were evaluated through IHC stains using SP1 monoclonal antibody for ER, and 1E2 monoclonal antibody for PR. All ER and PR immunostained slides were scanned, and invasive tumor areas were marked. Using the QuantCenter image analyzer provided by 3DHISTECH, IHC staining of hormone receptors was measured and converted to histochemical scores (H scores). Pearson correlation coefficients were calculated between Oncotype Dx hormone receptor scores and H scores, and between Oncotype Dx scores and Allred scores.
Results
H scores measured by an automated imaging system showed high concordance with RT-qPCR scores. ER concordance was 98.9% (92/93), and PR concordance was 91.4% (85/93). The correlation magnitude between automated H scores and RT-qPCR scores was high and comparable to those of Allred scores (for ER, 0.51 vs. 0.37 [p=.121], for PR, 0.70 vs. 0.72 [p=.39]).
Conclusions
Automated H scores showed a high concordance with quantitative mRNA expression levels measured by RT-qPCR.
Keyword
Figure
Reference
-
References
1. Umemura S, Kurosumi M, Moriya T, et al. Immunohistochemical evaluation for hormone receptors in breast cancer: a practically useful evaluation system and handling protocol. Breast Cancer. 2006; 13:232.
Article2. Hammond ME, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version). Arch Pathol Lab Med. 2010; 134:e48–72.3. Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004; 351:2817–26.
Article4. Xin L, Liu YH, Martin TA, Jiang WG. The era of multigene panels comes? The clinical utility of Oncotype DX and MammaPrint. World J Oncol. 2017; 8:34–40.
Article5. O’Connor SM, Beriwal S, Dabbs DJ, Bhargava R. Concordance between semiquantitative immunohistochemical assay and oncotype DX RT-PCR assay for estrogen and progesterone receptors. Appl Immunohistochem Mol Morphol. 2010; 18:268–72.6. Badve SS, Baehner FL, Gray RP, et al. Estrogen- and progesterone-receptor status in ECOG 2197: comparison of immunohistochemistry by local and central laboratories and quantitative reverse transcription polymerase chain reaction by central laboratory. J Clin Oncol. 2008; 26:2473–81.
Article7. Kraus JA, Dabbs DJ, Beriwal S, Bhargava R. Semi-quantitative immunohistochemical assay versus oncotype DX((R)) qRT-PCR assay for estrogen and progesterone receptors: an independent quality assurance study. Mod Pathol. 2012; 25:869–76.8. Bradshaw SH, Pidutti D, Gravel DH, Song X, Marginean EC, Robertson SJ. Predicting OncoDx recurrence scores with immunohistochemical markers. Appl Immunohistochem Mol Morphol. 2013; 21:490–6.
Article9. Park MM, Ebel JJ, Zhao W, Zynger DL. ER and PR immunohistochemistry and HER2 FISH versus oncotype DX: implications for breast cancer treatment. Breast J. 2014; 20:37–45.
Article10. Khoury T, Yan L, Liu S, Bshara W. Oncotype DX RT-qPCR assay for ER and PR correlation with IHC: a study of 3 different clones. Appl Immunohistochem Mol Morphol. 2015; 23:178–87.11. Hanna MG, Bleiweiss IJ, Nayak A, Jaffer S. Correlation of Oncotype DX recurrence score with histomorphology and immunohistochemistry in over 500 patients. Int J Breast Cancer. 2017; 2017:1257078.
Article12. Collins LC, Botero ML, Schnitt SJ. Bimodal frequency distribution of estrogen receptor immunohistochemical staining results in breast cancer: an analysis of 825 cases. Am J Clin Pathol. 2005; 123:16–20.13. Qureshi A, Pervez S. Allred scoring for ER reporting and it’s impact in clearly distinguishing ER negative from ER positive breast cancers. J Pak Med Assoc. 2010; 60:350–3.14. Allred DC, Mohsin SK. ER expression is not bimodal in breast cancer. Am J Clin Pathol. 2005; 124:474–5.15. Shousha S. Oestrogen receptor status of breast carcinoma: Allred/H score conversion table. Histopathology. 2008; 53:346–7.
Article16. Madabhushi A, Lee G. Image analysis and machine learning in digital pathology: challenges and opportunities. Med Image Anal. 2016; 33:170–5.
Article17. Veta M, Pluim JP, van Diest PJ, Viergever MA. Breast cancer histopathology image analysis: a review. IEEE Trans Biomed Eng. 2014; 61:1400–11.
Article18. Mohammed ZM, Edwards J, Orange C, et al. Breast cancer outcomes by steroid hormone receptor status assessed visually and by computer image analysis. Histopathology. 2012; 61:283–92.
Article19. Reisenbichler ES, Lester SC, Richardson AL, Dillon DA, Ly A, Brock JE. Interobserver concordance in implementing the 2010 ASCO/CAP recommendations for reporting ER in breast carcinomas: a demonstration of the difficulties of consistently reporting low levels of ER expression by manual quantification. Am J Clin Pathol. 2013; 140:487–94.20. Nassar A, Cohen C, Agersborg SS, et al. A new immunohistochemical ER/PR image analysis system: a multisite performance study. Appl Immunohistochem Mol Morphol. 2011; 19:195–202.21. Nadji M, Gomez-Fernandez C, Ganjei-Azar P, Morales AR. Immunohistochemistry of estrogen and progesterone receptors reconsidered: experience with 5,993 breast cancers. Am J Clin Pathol. 2005; 123:21–7.22. Iwamoto T, Booser D, Valero V, et al. Estrogen receptor (ER) mRNA and ER-related gene expression in breast cancers that are 1% to 10% ER-positive by immunohistochemistry. J Clin Oncol. 2012; 30:729–34.
Article23. Troxell ML, Long T, Hornick JL, Ambaye AB, Jensen KC. Comparison of estrogen and progesterone receptor antibody reagents using proficiency testing data. Arch Pathol Lab Med. 2017; 141:1402–12.
Article24. Zhang H, Han M, Varma KR, Clark BZ, Bhargava R, Dabbs DJ. High fidelity of breast biomarker metrics: a 10-year experience in a single, large academic institution. Appl Immunohistochem Mol Morphol. 2018; 26:697–700.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Expression of the pS2 Protein and Its Relation with Estrogen and Progesterone Receptor in Breast Cancer
- Immunohistochemical studies on the rate of positive reaction of estrogen receptors and progesterone receptors in the breast diseases
- A Study of Estrogen and Progesterone Receptor Expression in Neuroepithelial Tumors
- NanoString nCounter® Approach in Breast Cancer: A Comparative Analysis with Quantitative Real-Time Polymerase Chain Reaction, In Situ Hybridization, and Immunohistochemistry
- Estrogen receptor-α, progesterone receptor, and c-erbB/HER-family receptor mRNA detection and phenotype analysis in spontaneous canine models of breast cancer