J Bacteriol Virol.
2020 Jun;50(2):97-106. 10.4167/jbv.2020.50.2.097.
Effects of Exogenous N-Acyl-Homoserine Lactones on Biofilm Formation and Motility in Acinetobacter nosocomialis
- Affiliations
-
- 1Department of Microbiology and Medical Science, Chungnam National University School of Medicine, Daejeon 35015, Republic of Korea
- 2Department of Nanobiomedical Science, Dankook University, Cheonan 31116, Republic of Korea
- KMID: 2504385
- DOI: http://doi.org/10.4167/jbv.2020.50.2.097
Abstract
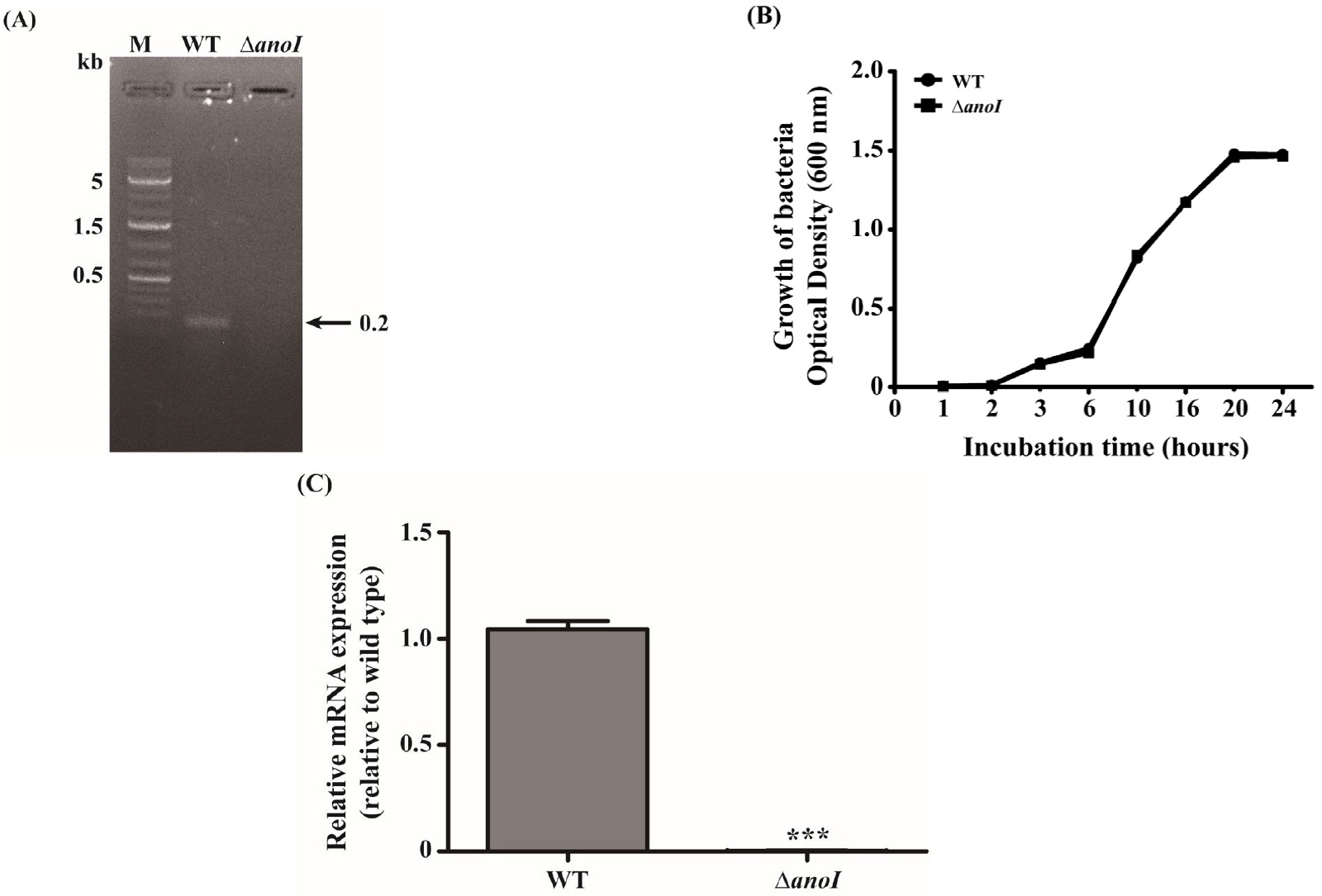
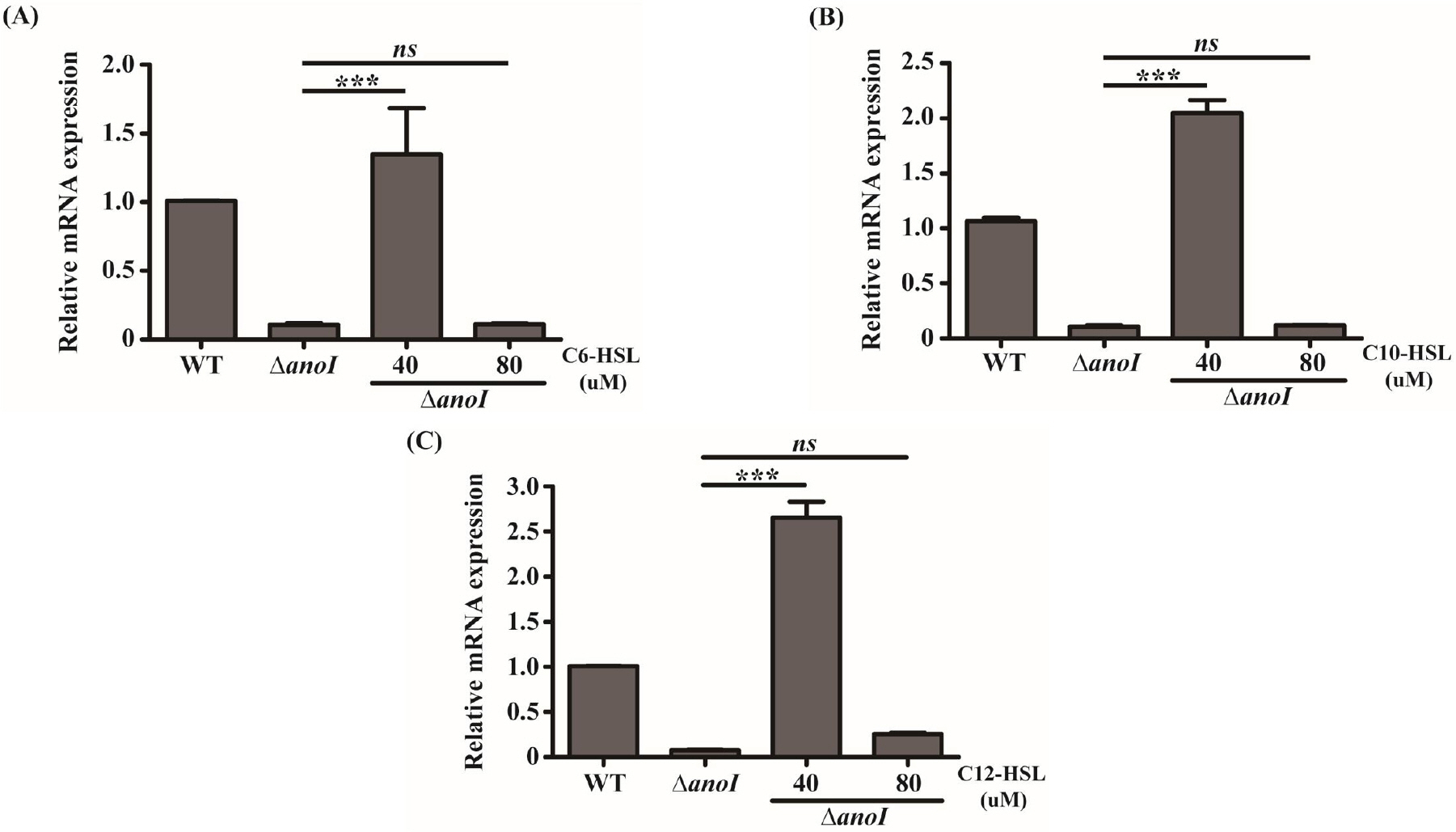
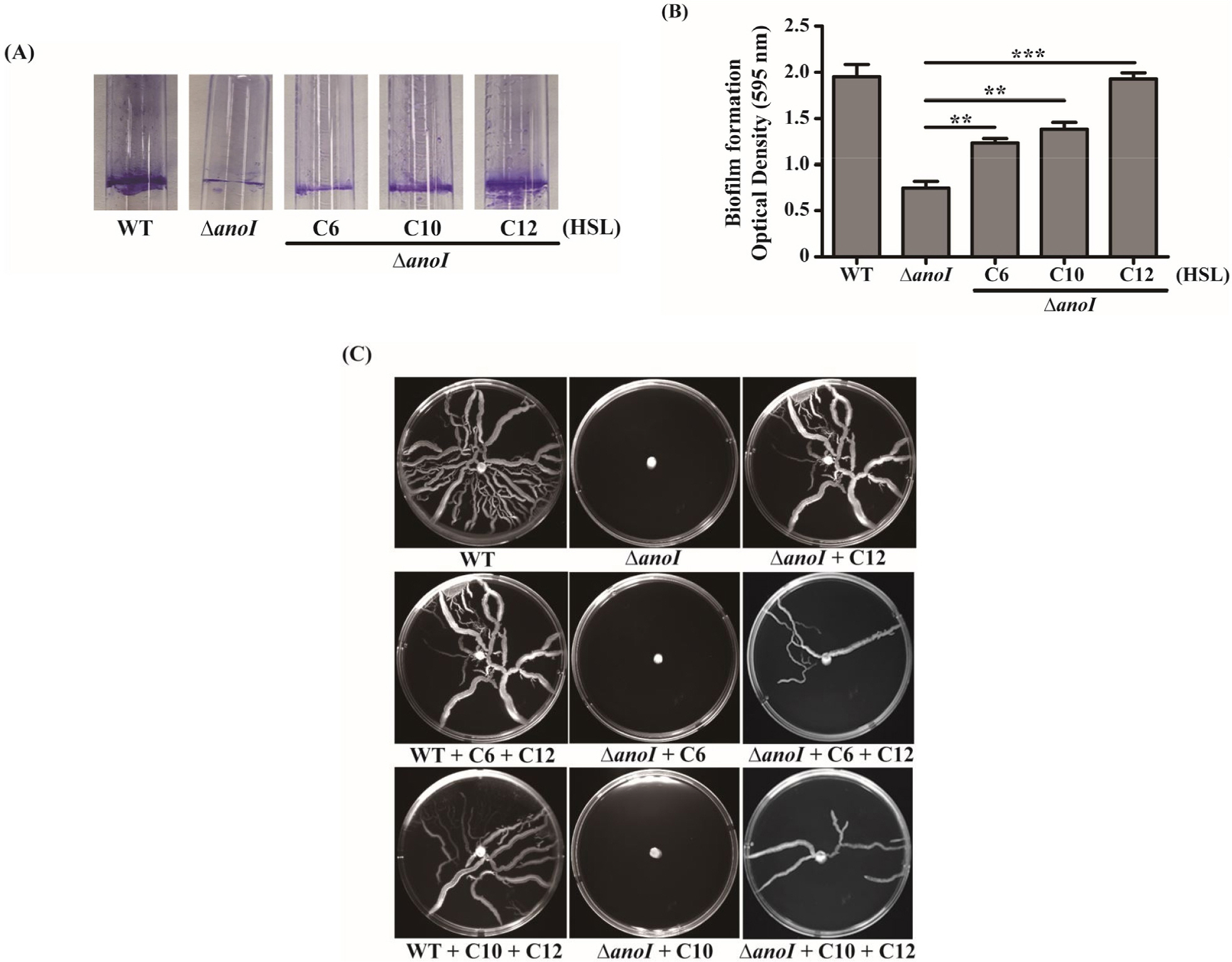
- One of the major factors contributing to drug resistance in Acinetobacter nosocomialis infections is biofilm development, which is facilitate by quorum-sensing (QS) systems. Quorum sensing by the LuxI and LuxR homologues, AnoI and AnoR, in A. nosocomialis plays a role in biofilm formation and motility of this pathogenic bacterium. The aim of this study was to evaluate the effects of exogenous N-acyl-homoserine lactones (AHLs) on the regulation of biofilm and motility of A. nosocomialis and anoI-deletion mutant. We found that anoR mRNA expression levels in the anoI-deletion mutant were increased in the presence of different types of AHLs compared with that in the absence of exogenous AHL. Among AHLs, C12-HSL appeared to exert the greatest stimulatory effect on biofilm formation and motility. Notably, the anoI-deletion mutant also exhibited significant decreases in expression of the biofilm- and motility-related genes, csuC, csuD and pilT, decreases that were attenuated by addition of exogenous AHLs. Combining the AHL C12-HSL with C6-HSL or C10-HSL exerted synergistic effects that restored the motility phenotype in the anoI-deletion mutant. Taken together, our data demonstrate that C12-HSL may act as an important signaling molecule in A. nosocomialis through regulation of biofilm formation and cell motility, potentially providing a new target for the control of A. nosocomialis infections.
Figure
Reference
-
1. Qi C, Malczynski M, Parker M, Scheetz MH. Characterization of genetic diversity of carbapenem-resistant Acinetobacter baumannii clinical strains collected from 2004 to 2007. J Clin Microbiol 2008;46:1106-9.DOI: 10.1128/JCM.01877-07. PMID: 18216212. PMCID: PMC2268351.2. Wang X, Chen T, Yu R, Lü X, Zong Z. Acinetobacter pittii and Acinetobacter nosocomialis among clinical isolates of the Acinetobacter calcoaceticus-baumannii complex in Sichuan, China. Diagn Microbiol Infect Dis 2013;76:392-5.DOI: 10.1016/j.diagmicrobio.2013.03.020. PMID: 23639796.3. Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 2008;21:538-82.DOI: 10.1128/CMR.00058-07. PMID: 18625687. PMCID: PMC2493088.4. Withers H, Swift S, Williams P. Quorum sensing as an integral component of gene regulatory networks in Gram-negative bacteria. Curr Opin Microbiol 2001;4:186-93.DOI: 10.1016/S1369-5274(00)00187-9.5. Bassler BL, Wright M, Showalter RE, Silverman MR. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol 1993;9:773-86.DOI: 10.1111/j.1365-2958.1993.tb01737.x. PMID: 8231809.6. Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol 2001;55:165-99.DOI: 10.1146/annurev.micro.55.1.165. PMID: 11544353.7. Oh MH, Choi CH. Role of LuxIR Homologue AnoIR in Acinetobacter nosocomialis and the Effect of Virstatin on the Expression of anoR Gene. J Microbiol Biotechnol 2015;25:1390-400.DOI: 10.4014/jmb.1504.04069. PMID: 25975610.8. Zhu YL, Hou HM, Zhang GL, Wang YF, Hao HS. AHLs Regulate Biofilm Formation and Swimming Motility of Hafnia alvei H4. Front Microbiol 2019;10:1330.DOI: 10.3389/fmicb.2019.01330. PMID: 31275267. PMCID: PMC6593095.9. Lee HW, Koh YM, Kim J, Lee JC, Lee YC, Seol SY, et al. Capacity of multidrug-resistant clinical isolates of Acinetobacter baumannii to form biofilm and adhere to epithelial cell surfaces. Clin Microbiol Infect 2008;14:49-54.DOI: 10.1111/j.1469-0691.2007.01842.x. PMID: 18005176.10. Armbruster CR, Parsek MR. New insight into the early stages of biofilm formation. Proc Natl Acad Sci USA 2018;115:4317-9.DOI: 10.1073/pnas.1804084115. PMID: 29632199. PMCID: PMC5924939.11. Tolker-Nielsen T. Biofilm Development. Microbiol Spectr 2015;3:MB-0001-2014.DOI: 10.1128/microbiolspec.MB-0001-2014. PMID: 26104692.12. Dunne WM Jr. Bacterial adhesion: seen any good biofilms lately? Clin Microbiol Rev 2002;15:155-66.DOI: 10.1128/CMR.15.2.155-166.2002. PMID: 11932228. PMCID: PMC118072.13. Fux CA, Costerton JW, Stewart PS, Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol 2005;13:34-40.DOI: 10.1016/j.tim.2004.11.010. PMID: 15639630.14. Nemec A, Krizova L, Maixnerova M, van der Reijden TJ, Deschaght P, Passet V, et al. Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU). Res Microbiol 2011;162:393-404.DOI: 10.1016/j.resmic.2011.02.006. PMID: 21320596.15. Oh MH, Lee JC, Kim J, Choi CH, Han K. Simple Method for Markerless Gene Deletion in Multidrug-Resistant Acinetobacter baumannii. Appl Environ Microbiol 2015;81:3357-68.DOI: 10.1128/AEM.03975-14. PMID: 25746991. PMCID: PMC4407223.16. Eberl L. N-acyl homoserinelactone-mediated gene regulation in gram-negative bacteria. Syst Appl Microbiol 1999;22:493-506.DOI: 10.1016/S0723-2020(99)80001-0.17. Saipriya K, Swathi CH, Ratnakar KS, Sritharan V. Quorum-sensing system in Acinetobacter baumannii: a potential target for new drug development. J Appl Microbiol 2020;128:15-27.DOI: 10.1111/jam.14330. PMID: 31102552.18. Rasmussen TB, Givskov M. Quorum sensing inhibitors: a bargain of effects. Microbiology 2006;152:895-904.DOI: 10.1099/mic.0.28601-0. PMID: 16549654.19. Li Z, Nair SK. Quorum sensing: how bacteria can coordinate activity and synchronize their response to external signals? Protein Sci 2012;21:1403-17.DOI: 10.1002/pro.2132. PMID: 22825856. PMCID: PMC3526984.20. Rémy B, Mion S, Plener L, Elias M, Chabrière E, Daudé D. Interference in Bacterial Quorum Sensing: A Biopharmaceutical Perspective. Front Pharmacol 2018;9:203.DOI: 10.3389/fphar.2018.00203. PMID: 29563876. PMCID: PMC5845960.21. Gurich N, González JE. Role of quorum sensing in Sinorhizobium meliloti-Alfalfa symbiosis. J Bacteriol 2009;191:4372-82.DOI: 10.1128/JB.00376-09. PMID: 19395488. PMCID: PMC2698488.22. Andersson RA, Eriksson AR, Heikinheimo R, Mäe A, Pirhonen M, Kõiv V, et al. Quorum sensing in the plant pathogen Erwinia carotovora subsp. carotovora: the role of expR (Ecc). Mol Plant Microbe Interact 2000;13:384-93.DOI: 10.1094/MPMI.2000.13.4.384. PMID: 10755301.23. Nasser W, Bouillant ML, Salmond G, Reverchon S. Characterization of the Erwinia chrysanthemi expI-expR locus directing the synthesis of two N-acyl-homoserine lactone signal molecules. Mol Microbiol 1998;29:1391-405.DOI: 10.1046/j.1365-2958.1998.01022.x. PMID: 9781877.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- In vitro Comparison of Anti-Biofilm Effects against Carbapenem-Resistant Acinetobacter baumannii: Imipenem, Colistin, Tigecycline, Rifampicin and Combinations
- Association of biofilm production with colonization among clinical isolates of Acinetobacter baumannii
- Octanoic acid-rich diet alleviates breast cancerinduced bone pain via the acyl-ghrelin/NPY pathway
- Inhibitory Effect of Pentose on Biofilm Formation by Oral Bacteria
- Inhibition of biofilm formation of periodontal pathogens by D-Arabinose