J Bacteriol Virol.
2020 Jun;50(2):65-75. 10.4167/jbv.2020.50.2.065.
Strength and Weakness of Molecular Identification Strategies Against Causative Viral Agent from Emerging COVID-19
- Affiliations
-
- 1Department of Microbiology, Institute for Viral Diseases, College of Medicine, Korea University, 73, Goryeodae-ro, Seongbuk-gu, Seoul 02841,Republic of Korea
- KMID: 2504383
- DOI: http://doi.org/10.4167/jbv.2020.50.2.065
Abstract
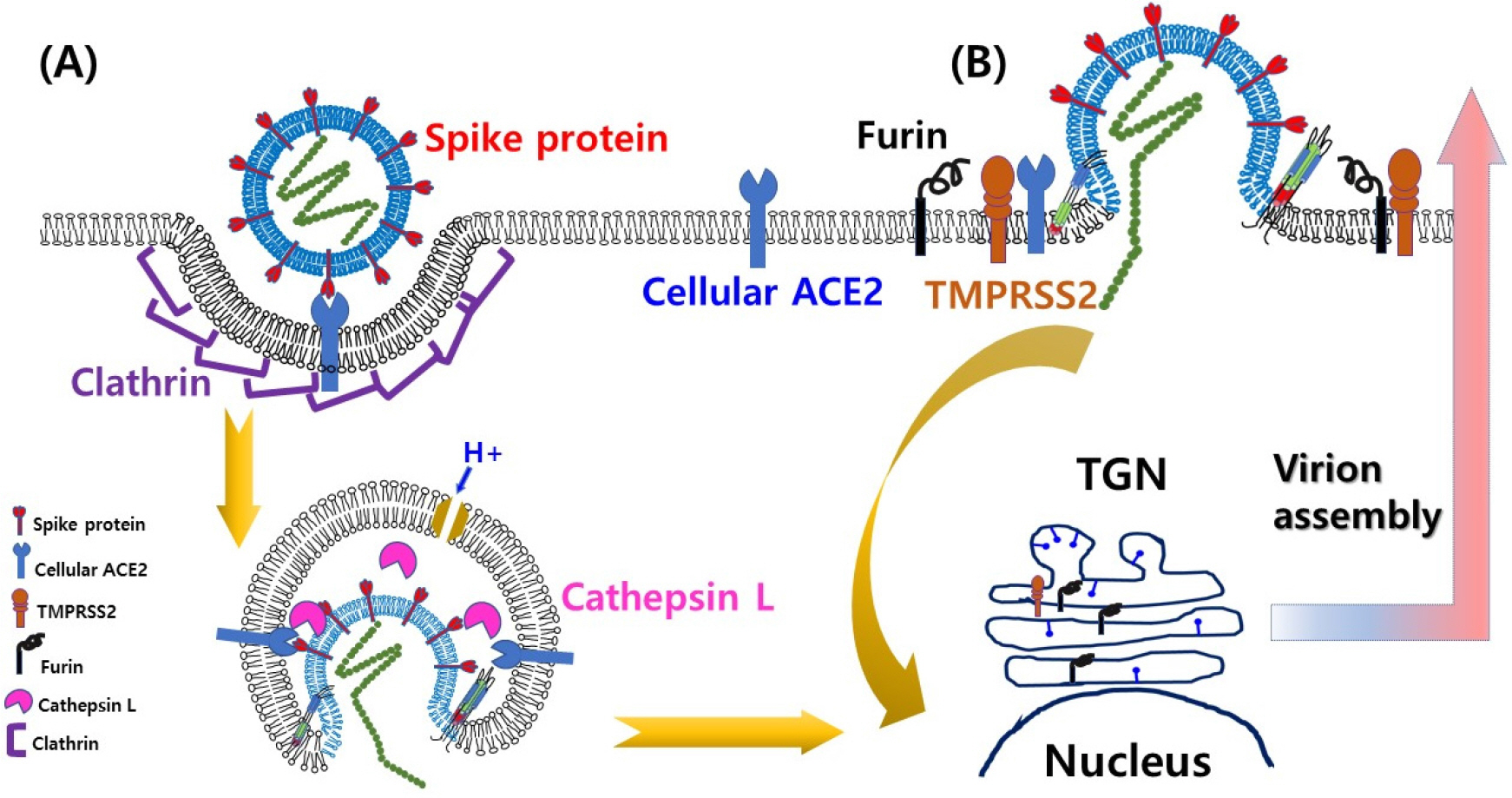
- A century ago, more exactly 102 years ago, there was a devastating pandemic of influenza in 1918 and thereafter, periodic recurrences of pandemic events have been reported in the human population. Unfortunately, whenever it happened, the outcome was concomitant with over millions of death tolls due to considerably higher case fatality rates, compared to other infectious diseases at that time. In this regard, pandemics, which continued at irregular time intervals, give a great significance to global public health responses. However, it is far from feasibility to predict when a next pandemic will begin and how much disease burden will be despite our efforts to utilize all kinds of available scientific information and knowledge. The one clear thing is that approximately 70% of the causative agents of emerging and/or re-emerging diseases including COVID-19 (coronavirus disease 2019), which has been started from Wuhan province, China in December 2019 and has resulted in more than 4 million human cases within a few months, are viruses. Therefore, it is very important to secure fast and accurate identification methods of a causative pathogen in order to provide scientific clues and to prepare in advance for the abrupt occurrence of unknown viral diseases in a timely manner. In this review, the current status and future perspectives of the molecular technology for identification of viral pathogens such as SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) with regard to rapid public health responses in the early stage of infectious diseases including COVID-19, will be discussed.
Keyword
Figure
Reference
-
1. WORLDOMETER https://www.worldometers.info/coronavirus/.2. Korea Centers for Disease Control and Prevention (KCDC). The updates on COVID-19 in Korea as of 14 June.3. Amanat F, Krammer F. SARS-CoV-2 Vaccines: Status Report. Immunity 2020; 52:583-9.DOI: 10.1016/j.immuni.2020.03.007. PMID: 32259480. PMCID: PMC7136867.4. Li H, Zhou Y, Zhang M, Wang H, Zhao Q, Liu J. Updated Approaches against SARS-CoV-2. Antimicrob Agents Chemother 2020;64:e00483-20.DOI: 10.1128/AAC.00483-20. PMID: 32205349. PMCID: PMC7269512.5. Hunter P. Viral vigilance. New surveillance strategies and methods help to identify dangerous pathogens earlier: a prerequisite for efficient countermeasures. EMBO 2008;9:948-50.DOI: 10.1038/embor.2008.181. PMID: 18830223. PMCID: PMC2572123.6. Zhang N, Li C, Hu Y, Li K, Liang J, Wang L, et al. Current development of COVID-19 diagnostics, vaccines and therapeutics. Microbes Infect 2020;S1286-4579(20)30079-4.7. Ahn DG, Shin HJ, Kim MH, Lee S, Kim HS, Myoung J, et al. Current Status of Epidemiology, Diagnosis, Therapeutics, and Vaccines for Novel Coronavirus Disease 2019 (COVID-19). J Microbiol Biotechnol 2020;30:313-24.DOI: 10.4014/jmb.2003.03011. PMID: 32238757.8. Tu YF, Chien CS, Yarmishyn AA, Lin YY, Luo YH, Lin YT, et al. A Review of SARS-CoV-2 and the Ongoing Clinical Trials. Int J Mol Sci 2020;21:2657.DOI: 10.3390/ijms21072657. PMID: 32290293. PMCID: PMC7177898.9. Ciliberto G, Mancini R, Paggi MG. Drug repurposing against COVID-19: focus on anticancer agents. J Exp Clin Cancer Res 2020;39:86.DOI: 10.1186/s13046-020-01590-2. PMID: 32398164. PMCID: PMC7214852.10. Ma C, Su S, Wang J, Wei L, Du L, Jiang S. From SARS-CoV to SARS-CoV-2: safety and broad-spectrum are important for coronavirus vaccine development. Microbes Infect 2020:S1286-4579(20)30082-4.11. Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol 2020;38:1-9.12. de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol 2016;14:523-34.DOI: 10.1038/nrmicro.2016.81. PMID: 27344959. PMCID: PMC7097822.13. Liu J, Zheng X, Tong Q, Li W, Wang B, Sutter K, et al. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J Med Virol 2020;92:491-4.DOI: 10.1002/jmv.25709. PMID: 32056249. PMCID: PMC7166760.14. Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 2009;459:931-9.DOI: 10.1038/nature08157. PMID: 19525932. PMCID: PMC2873852.15. Hematian A, Sadeghifard N, Mohebi R, Taherikalani M, Nasrolahi A, Amraei M, et al. Traditional and Modern Cell Culture in Virus Diagnosis. Osong Public Health Res Perspect 2016;7:77-82.DOI: 10.1016/j.phrp.2015.11.011. PMID: 27169004. PMCID: PMC4850366.16. Leland DS, Ginocchio CC. Role of cell culture for virus detection in the age of technology. Clin Microbiol Rev 2007;20:49-78.DOI: 10.1128/CMR.00002-06. PMID: 17223623. PMCID: PMC1797634.17. Matsuyama S, Nao N, Shirato K, Kawase M, Saito S, Takayama I. et al., Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A 2020;117:7001-3.DOI: 10.1073/pnas.2002589117. PMID: 32165541. PMCID: PMC7132130.18. Ma D, Chen CB, Jhanji V, Xu C, Yuan XL, Liang JJ, et al. Expression of SARS-CoV-2 receptor ACE2 and TMPRSS2 in human primary conjunctival and pterygium cell lines and in mouse cornea. Eye (Lond) 2020:1-8.DOI: 10.1038/s41433-020-0939-4. PMID: 32382146. PMCID: PMC7205026.19. Guo Y, Tisoncik J, McReynolds S, Farzan M, Prabhakar BS, Gallagher T, et al. Identification of a new region of SARS-CoV S protein critical for viral entry. J Mol Biol 2009;394:600-5.DOI: 10.1016/j.jmb.2009.10.032. PMID: 19853613. PMCID: PMC2794126.20. Leland DS, Ginocchio CC. Role of Cell Culture for Virus Detection in the Age of Technology. Clin Microbiol Rev 2007;20:49-78.DOI: 10.1128/CMR.00002-06. PMID: 17223623. PMCID: PMC1797634.21. Fernandez-Garcia MD, Simon-Loriere E, Kebe O, Sakuntabhai A, Ndiaye K. Identification and molecular characterization of the first complete genome sequence of Human Parechovirus type 15. Sci Rep 2020;10:6759.DOI: 10.1038/s41598-020-63467-w. PMID: 32317760. PMCID: PMC7174385.22. Montmayeur AM, Ng TF, Schmidt A, Zhao K, Magaña L, Iber J, et al. High-Throughput Next-Generation Sequencing of Polioviruses. J Clin Microbiol 2017;55:606-15.DOI: 10.1128/JCM.02121-16. PMID: 27927929. PMCID: PMC5277531.23. Klenner J, Kohl C, Dabrowski PW, Nitsche A. Comparing Viral Metagenomic Extraction Methods. Curr Issues Mol Biol 2017;24:59-70.DOI: 10.21775/cimb.024.059. PMID: 28686568.24. Roth WK, Seifried E. Yield and future issues of nucleic acid testing. Transfus Clin Biol 2001;8:282-4.DOI: 10.1016/S1246-7820(01)00115-X.25. Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature 2020;579:265-9.DOI: 10.1038/s41586-020-2008-3. PMID: 32015508. PMCID: PMC7094943.26. Wang C, Liu Z, Chen Z, Huang X, Xu M, He T, et al. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J Med Virol 2020;92:667-74.DOI: 10.1002/jmv.25762. PMID: 32167180. PMCID: PMC7228400.27. Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020;395:565-74.DOI: 10.1016/S0140-6736(20)30251-8.28. Kim JM, Chung YS, Jo HJ, Lee NJ, Kim MS, Woo SH, et al. Identification of Coronavirus Isolated from a Patient in Korea with COVID-19. Osong Public Health Res Perspect 2020;11:3-7.DOI: 10.24171/j.phrp.2020.11.1.02. PMID: 32149036. PMCID: PMC7045880.29. Roingeard P, Raynal PI, Eymieux S, Blanchard E. Virus detection by transmission electron microscopy: Still useful for diagnosis and a plus for biosafety. Rev Med Virol 2019;29:e2019.DOI: 10.1002/rmv.2019. PMID: 30411832. PMCID: PMC7169071.30. Gullett JC, Nolte FS. Quantitative nucleic acid amplification methods for viral infections. Clin Chem 2015;61:72-8.DOI: 10.1373/clinchem.2014.223289. PMID: 25403817.31. Merckx J, Wali R, Schiller I, Caya C, Gore GC, Chartrand C, et al. Diagnostic Accuracy of Novel and Traditional Rapid Tests for Influenza Infection Compared With Reverse Transcriptase Polymerase Chain Reaction: A Systematic Review and Meta-analysis. Ann Intern Med 2017;167:394-409.DOI: 10.7326/M17-0848. PMID: 28869986.32. Mahony JB, Petrich A, Smieja M. Molecular diagnosis of respiratory virus infections. Crit Rev Clin Lab Sci 2011;48:217-49.DOI: 10.3109/10408363.2011.640976. PMID: 22185616.33. Waters DL, Shapter FM. The polymerase chain reaction (PCR): general methods. Methods Mol Biol 2014;1099:65-75.DOI: 10.1007/978-1-62703-715-0_7. PMID: 24243196.34. Lorenz TC. Polymerase chain reaction: basic protocol plus troubleshooting and optimization strategies. J Vis Exp 2012;22:e3998.DOI: 10.3791/3998. PMID: 22664923. PMCID: PMC4846334.35. Rogers BB. The Evolution of the Polymerase Chain Reaction to Diagnose Childhood Infections. Pediatr Dev Pathol 2015;18:495-503.DOI: 10.2350/15-05-1643-OA.1. PMID: 26701384.36. Richards GP. Limitations of molecular biological techniques for assessing the virological safety of foods. J Food Prot 1999;62:691-7.DOI: 10.4315/0362-028X-62.6.691. PMID: 10382664.37. Elnifro EM, Ashshi AM, Cooper RJ, Klapper PE. Multiplex PCR: optimization and application in diagnostic virology. Clin Microbiol Rev 2000;13:559-70.DOI: 10.1128/CMR.13.4.559. PMID: 11023957. PMCID: PMC88949.38. Bacich DJ, Sobek KM, Cummings JL, Atwood AA, O'Keefe DS. False negative results from using common PCR reagents. BMC Res Notes 2011;4:457.DOI: 10.1186/1756-0500-4-457. PMID: 22032271. PMCID: PMC3219698.39. Burkardt HJ. Standardization and quality control of PCR analyses. Clin Chem Lab Med 2000;38:87-91.DOI: 10.1515/CCLM.2000.014. PMID: 10834394.40. Sánchez-Seco MP, Rosario D, Hernández L, Domingo C, Valdés K, Guzmán MG, et al. Detection and subtyping of dengue 1-4 and yellow fever viruses by means of a multiplex RT-nested-PCR using degenerated primers. Trop Med Int Health 2006;11:1432-41.DOI: 10.1111/j.1365-3156.2006.01696.x. PMID: 16930266.41. Kim JY, Choe PG, Oh Y, Oh KJ, Kim J, Park SJ, et al. The First Case of 2019 Novel Coronavirus Pneumonia Imported into Korea from Wuhan, China: Implication for Infection Prevention and Control Measures. J Korean Med Sci 2020;35:e61.DOI: 10.3346/jkms.2020.35.e61. PMID: 32030925. PMCID: PMC7008073.42. van Kasteren PB, van der Veer B, van den Brink S, Wijsman L, de Jonge J, van den Brandt A, et al. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J Clin Virol 2020;128:104412.DOI: 10.1016/j.jcv.2020.104412. PMID: 32416600. PMCID: PMC7206434.43. Vogels CBF, Brito AF, Wyllie AL, Fauver JR, Ott IM, Kalinich CC, et al. Analytical sensitivity and efficiency comparisons of SARS-COV-2 qRT-PCR primer-probe sets. medRxiv 2020.44. Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain reaction product by utilizing the 5'----3' exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci U S A 1991;88:7276-80.DOI: 10.1073/pnas.88.16.7276. PMID: 1871133. PMCID: PMC52277.45. Manganelli R, Tyagi S, Smith I. Real Time PCR Using Molecular Beacons: A New Tool to Identify Point Mutations and to Analyze Gene Expression in Mycobacterium tuberculosis. Methods Mol Med 2001;54:295-310.DOI: 10.1385/1-59259-147-7:295. PMID: 21341083.46. Ponchel F, Toomes C, Bransfield K, Leong FT, Douglas SH, Field SL, et al. Real-time PCR based on SYBR-Green I fluorescence: An alternative to the TaqMan assay for a relative quantification of gene rearrangements, gene amplifications and micro gene deletions. BMC Biotechnol 2003;3:18.DOI: 10.1186/1472-6750-3-18. PMID: 14552656. PMCID: PMC270040.47. Korea Centers for Disease Control and Prevention (KCDC). Approval for emergency use of test reagent for COVID-19 gene detection (2020). https://www.cdc.go.kr/board/board.es?mid=a20505000000&bid=0017.48. Carlson CS, Emerson RO, Sherwood AM, Desmarais C, Chung MW, Parsons JM, et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun 2013;4:2680.DOI: 10.1038/ncomms3680. PMID: 24157944.49. Jung S, Kim J, Lee DJ, Oh EH, Lim H, Kim KP, et al. Extensible Multiplex Real-time PCR of MicroRNA Using Microparticles. Sci Rep 2016;6:22975.DOI: 10.1038/srep22975. PMID: 26964639. PMCID: PMC4786821.50. Houldcroft CJ, Beale MA, Breuer J. Clinical and biological insights from viral genome sequencing. Nat Rev Microbiol 2017;15:183-92.DOI: 10.1038/nrmicro.2016.182. PMID: 28090077. PMCID: PMC7097211.51. Kim JH, Kang M, Park E, Chung DR, Kim J, Hwang ES. A Simple and Multiplex Loop-Mediated Isothermal Amplification (LAMP) Assay for Rapid Detection of SARS-CoV. Biochip J 2019;13:341-51.DOI: 10.1007/s13206-019-3404-3. PMID: 32226589. PMCID: PMC7097549.52. Li C, Ren L. Recent progress on the diagnosis of 2019 Novel Coronavirus. Transbound Emerg Dis 2020:10.DOI: 10.1111/tbed.13620. PMID: 32395897. PMCID: PMC7272792.53. Yan C, Cui J, Huang L, Du B, Chen L, Xue G, et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin Microbiol Infect 2020;26:773-9.DOI: 10.1016/j.cmi.2020.04.001. PMID: 32276116. PMCID: PMC7144850.54. Xiang X, Qian K, Zhang Z, Lin F, Xie Y, Liu Y, et al. CRISPR-Cas Systems Based Molecular Diagnostic Tool for Infectious Diseases and Emerging 2019 Novel Coronavirus (COVID-19) Pneumonia. J Drug Target 2020:1-5.DOI: 10.1080/1061186X.2020.1769637. PMID: 32401064. PMCID: PMC7265108.55. Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018;360:436-9.DOI: 10.1126/science.aar6245. PMID: 29449511. PMCID: PMC6628903.56. Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol 2020.DOI: 10.1038/s41587-020-0513-4. PMID: 32300245.57. Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020;181:914-21.DOI: 10.1016/j.cell.2020.04.011. PMID: 32330414. PMCID: PMC7179501.58. Fauver JR, Petrone ME, Hodcroft EB, Shioda K, Ehrlich HY, Watts AG, et al. Coast-to-Coast Spread of SARS-CoV-2 during the Early Epidemic in the United States Cell 2020;181:990-6.DOI: 10.1016/j.cell.2020.04.021. PMID: 32386545. PMCID: PMC7204677.59. Lu J, du Plessis L, Liu Z, Hill V, Kang M, Lin H, et al. Genomic Epidemiology of SARS-CoV-2 in Guangdong Province, China. Cell 2020;181:997-1003.DOI: 10.1016/j.cell.2020.04.023. PMID: 32359424. PMCID: PMC7192124.60. Ward AB, Wilson IA. Innovations in structure-based antigen design and immune monitoring for next generation vaccines. Curr Opin Immunol 2020;65:50-6.DOI: 10.1016/j.coi.2020.03.013. PMID: 32387642. PMCID: PMC7174181.61. Wang LF, Anderson DE, Mackenzie JS, Merson MH. From Hendra to Wuhan: what has been learned in responding to emerging zoonotic viruses. Lancet 2020;395:e33-4.DOI: 10.1016/S0140-6736(20)30350-0.62. Schobel SA, Stucker KM, Moore ML, Anderson LJ, Larkin EK, Shankar J, et al. Respiratory Syncytial Virus whole-genome sequencing identifies convergent evolution of sequence duplication in the C-terminus of the G gene. Sci Rep 2016;6:26311.DOI: 10.1038/srep26311. PMID: 27212633. PMCID: PMC4876326.63. Peserico A, Marcacci M, Malatesta D, Di Domenico M, Pratelli A, Mangone I, et al. Diagnosis and characterization of canine distemper virus through sequencing by MinION nanopore technology. Sci Rep 2019;9:1714.DOI: 10.1038/s41598-018-37497-4. PMID: 30737428. PMCID: PMC6368598.64. Lu H, Giordano F, Ning Z. Oxford Nanopore MinION Sequencing and Genome Assembly. Genomics Proteomics Bioinformatics 2016;14:265-79.DOI: 10.1016/j.gpb.2016.05.004. PMID: 27646134. PMCID: PMC5093776.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Post COVID-19 Emerging Infectious Diseases: What is the Next Pandemic Agent?
- Will Mutations in the Spike Protein of SARS-CoV-2 Lead to the Failure of COVID-19 Vaccines?
- Effectiveness of massage and range of motion exercises on muscle strength and intensive care unit-acquired weakness in Iranian patients with COVID-19: a randomized parallel-controlled trial
- Pulmonary Contusion Similar to COVID-19 Pneumonia
- The Management of Thyroid Disease in COVID-19 Pandemic