J Korean Med Sci.
2021 May;36(18):e124. 10.3346/jkms.2021.36.e124.
Will Mutations in the Spike Protein of SARS-CoV-2 Lead to the Failure of COVID-19 Vaccines?
- Affiliations
-
- 1Tuberculosis Prevention and Control Key Laboratory/Beijing Key Laboratory of New Techniques of Tuberculosis Diagnosis and Treatment, Institute for Tuberculosis Research, 8th Medical Center, Chinese PLA General Hospital, Beijing, China
- KMID: 2515857
- DOI: http://doi.org/10.3346/jkms.2021.36.e124
Abstract
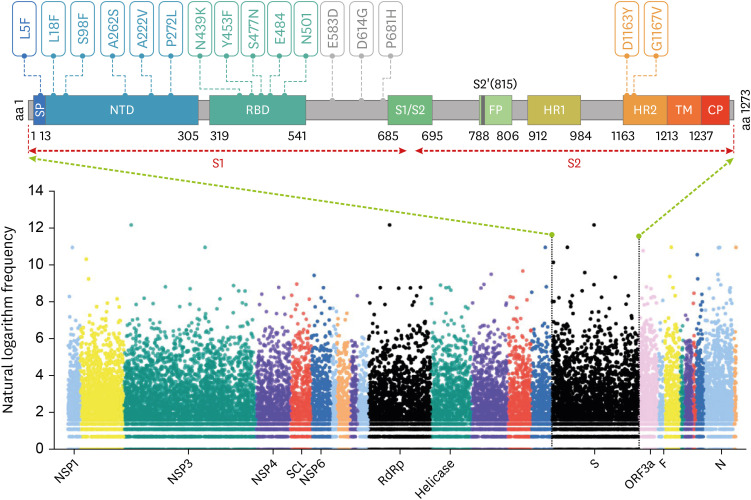
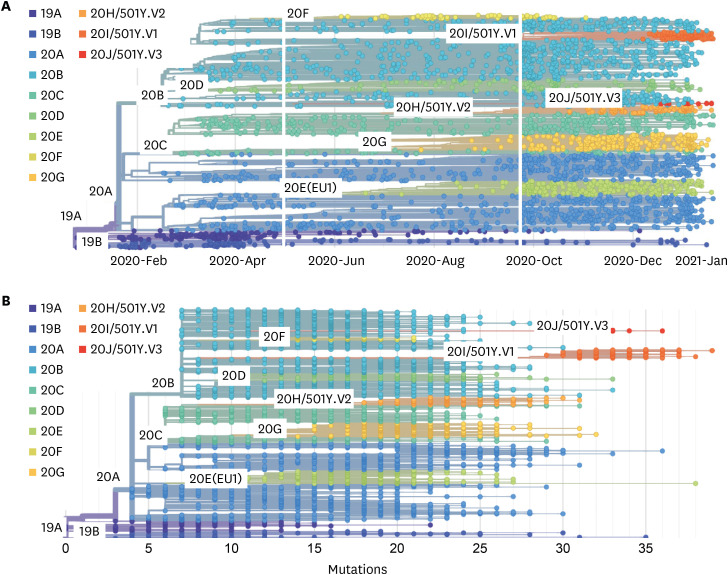
- Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is the causative agent of coronavirus disease 2019 (COVID-19), which has spread worldwide since it was first identified in Wuhan, China, at the end of 2019. With the global transmission of the virus, a large number of SARS-CoV-2 variants have also appeared, especially, emerging strains that have recently been discovered in the United Kingdom (variant 20I/501Y.V1, lineage B.1.1.7), South Africa (variant 20H/501Y.V2, lineage B.1.351), and Brazil (variant 20 J/501Y.V3, and lineage P.1). The common feature of these variants is that they share the N501Y mutation involving the SARS-CoV-2 spike (S) protein, which is precisely the target of most COVID-19 vaccines. Furthermore, mutations such as N501Y, E484K, and K417N in the S protein may affect viral fitness and transmissibility. However, current research on the impact of these variants on COVID-19 vaccines is still lacking. Herein, we briefly explain why most COVID-19 vaccines target the S protein, update the progress of research regarding S protein-related COVID-19 vaccines, review the latest studies concerning the effects of S protein variants on COVID-19 vaccines, and finally, propose certain strategies to deal with SARS-CoV-2 variants.
Keyword
Figure
Cited by 2 articles
-
Effectiveness and Safety of Regdanvimab in Patients With Mild-To-Moderate COVID-19: A Retrospective Cohort Study
Susin Park, Nam Kyung Je, Dong Wan Kim, Miran Park, Jeonghun Heo
J Korean Med Sci. 2022;37(13):e102. doi: 10.3346/jkms.2022.37.e102.The Higher Incidence of COVID-19 in Patients With Non-Tuberculous Mycobacterial Pulmonary Disease: A Single Center Experience in Korea
Sang Hyuk Kim, Byung Woo Jhun, Byeong-Ho Jeong, Hye Yun Park, Hojoong Kim, O Jung Kwon, Sun Hye Shin
J Korean Med Sci. 2022;37(32):e250. doi: 10.3346/jkms.2022.37.e250.
Reference
-
1. Oliver SE, Gargano JW, Marin M, Wallace M, Curran KG, Chamberland M, et al. The Advisory Committee on Immunization Practices' interim recommendation for use of Moderna COVID-19 vaccine - United States, December 2020. MMWR Morb Mortal Wkly Rep. 2021; 69(5152):1653–1656. PMID: 33382675.
Article2. Mahase E. Covid-19: UK approves Moderna vaccine to be given as two doses 28 days apart. BMJ. 2021; 372:n74. PMID: 33431500.
Article3. Mahase E. Covid-19: UK approves Pfizer and BioNTech vaccine with rollout due to start next week. BMJ. 2020; 371:m4714. PMID: 33268330.
Article4. Mahase E. Covid-19: Russia approves vaccine without large scale testing or published results. BMJ. 2020; 370:m3205. PMID: 32816758.
Article5. Voysey M, Costa Clemens SA, Madhi SA, Weckx LY, Folegatti PM, Aley PK, et al. Single-dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine: a pooled analysis of four randomised trials. Lancet. 2021; 397(10277):881–891. PMID: 33617777.6. Stephenson KE, Le Gars M, Sadoff J, de Groot AM, Heerwegh D, Truyers C, et al. Immunogenicity of the Ad26.COV2.S vaccine for COVID-19. JAMA. Forthcoming. 2021; DOI: 10.1001/jama.2021.3645.7. Galloway SE, Paul P, MacCannell DR, Johansson MA, Brooks JT, MacNeil A, et al. Emergence of SARS-CoV-2 B.1.1.7 lineage - United States, December 29, 2020–January 12, 2021. MMWR Morb Mortal Wkly Rep. 2021; 70(3):95–99. PMID: 33476315.
Article8. Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. Forthcoming. 2020; DOI: 10.1101/2020.12.21.20248640.
Article9. Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020; 367(6483):1260–1263. PMID: 32075877.
Article10. Duan L, Zheng Q, Zhang H, Niu Y, Lou Y, Wang H. The SARS-CoV-2 spike glycoprotein biosynthesis, structure, function, and antigenicity: implications for the design of spike-based vaccine immunogens. Front Immunol. 2020; 11:576622. PMID: 33117378.
Article11. Rui R, Tian M, Tang ML, Ho GT, Wu CH. Analysis of the spread of COVID-19 in the USA with a spatio-temporal multivariate time series model. Int J Environ Res Public Health. 2021; 18(2):774.
Article12. Naveca F, Nascimento V, Souza V, Corado A, Nascimento F, Silva G, et al. Phylogenetic relationship of SARS-CoV-2 sequences from Amazonas with emerging Brazilian variants harboring mutations E484K and N501Y in the spike protein. Updated 2020. Accessed March 19, 2021. https://virological.org/t/phylogenetic-relationship-of-sars-cov-2-sequences-from-amazonas-with-emerging-brazilian-variants-harboring-mutations-e484k-and-n501y-in-the-spike-protein/585.13. GISAID. UK reports new variant, termed VUI 202012/01. Updated 2020. Accessed March 19, 2021. https://www.gisaid.org/references/gisaid-in-the-news/uk-reports-new-variant-termed-vui-20201201/.14. Teruel N, Mailhot O, Najmanovich RJ. Modelling conformational state dynamics and its role on infection for SARS-CoV-2 Spike protein variants. bioRxiv. Forthcoming. 2020; DOI: 10.1101/2020.12.16.423118.
Article15. Rathnasinghe R, Jangra S, Cupic A, Martínez-Romero C, Mulder LCF, Kehrer T, et al. The N501Y mutation in SARS-CoV-2 spike leads to morbidity in obese and aged mice and is neutralized by convalescent and post-vaccination human sera. medRxiv. Forthcoming. 2021; DOI: 10.1101/2021.01.19.21249592.
Article16. Koyama T, Weeraratne D, Snowdon JL, Parida L. Emergence of drift variants that may affect COVID-19 vaccine development and antibody treatment. Pathogens. 2020; 9(5):324.
Article17. Weissman D, Alameh MG, de Silva T, Collini P, Hornsby H, Brown R, et al. D614G spike mutation increases SARS CoV-2 susceptibility to neutralization. Cell Host Microbe. 2021; 29(1):23–31.e4. PMID: 33306985.
Article18. Zhang L, Jackson CB, Mou H, Ojha A, Peng H, Quinlan BD, et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun. 2020; 11(1):6013. PMID: 33243994.
Article19. Chen J, Wang R, Wang M, Wei GW. Mutations strengthened SARS-CoV-2 infectivity. J Mol Biol. 2020; 432(19):5212–5226. PMID: 32710986.
Article20. Gaebler C, Wang Z, Lorenzi JCC, Muecksch F, Finkin S, Tokuyama M, et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021; 591(7851):639–644. PMID: 33461210.
Article21. Greaney AJ, Loes AN, Crawford KHD, Starr TN, Malone KD, Chu HY, et al. Comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human serum antibodies. Cell Host Microbe. 2021; 29(3):463–476.e6. PMID: 33592168.
Article22. Andreano E, Piccini G, Licastro D, Casalino L, Johnson NV, Paciello I, et al. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv. Forthcoming. 2020; DOI: 10.1101/2020.12.28.424451.
Article23. Wu K, Werner AP, Moliva JI, Koch M, Choi A, Stewart-Jones GBE, et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv. Forthcoming. 2021; DOI: 10.1101/2021.01.25.427948.
Article24. Wang Z, Schmidt F, Weisblum Y, Muecksch F, Barnes CO, Finkin S, et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. Forthcoming. 2021; DOI: 10.1038/s41586-021-03324-6.
Article25. Sapkal GN, Yadav PD, Ella R, Deshpande GR, Sahay RR, Gupta N, et al. Neutralization of UK-variant VUI-202012/01 with COVAXIN vaccinated human serum. bioRxiv. Forthcoming. 2021; DOI: 10.1101/2021.01.26.426986.
Article26. Emary KRW, Golubchik T, Aley PK, Ariani CV, Angus B, Bibi S, et al. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B.1.1.7): an exploratory analysis of a randomised controlled trial. Lancet. 2021; 397(10282):1351–1362. PMID: 33798499.27. van der Lubbe JEM, Rosendahl Huber SK, Vijayan A, Dekking L, van Huizen E, Vreugdenhil J, et al. Ad26.COV2.S protects Syrian hamsters against G614 spike variant SARS-CoV-2 and does not enhance respiratory disease. NPJ Vaccines. 2021; 6(1):39. PMID: 33741993.
Article28. Novavax. Novavax COVID-19 vaccine demonstrates 89.3% efficacy in UK phase 3 trial. Updated 2021. Accessed January 28, 2021. https://ir.novavax.com/news-releases/news-release-details/novavax-covid-19-vaccine-demonstrates-893-efficacy-uk-phase-3.29. Liu Z, VanBlargan LA, Bloyet LM, Rothlauf PW, Chen RE, Stumpf S, et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021; 29(3):477–488.e4. PMID: 33535027.
Article30. Pujhari S, Rasgon JL. Mice with humanized-lungs and immune system - an idealized model for COVID-19 and other respiratory illness. Virulence. 2020; 11(1):486–488. PMID: 32434416.
Article31. Hansen J, Baum A, Pascal KE, Russo V, Giordano S, Wloga E, et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020; 369(6506):1010–1014. PMID: 32540901.
Article32. Environmental and Modelling Group (EMG) and New and Emerging Respiratory Virus Threats Advisory Group (NERVTAG). SARS-COV-2: transmission routes and environments, 22 October 2020. Updated 2020. Accessed March 19, 2021. https://www.gov.uk/government/publications/sars-cov-2-transmission-routes-and-environments-22-october-2020.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Spike protein D614G and RdRp P323L: the SARS-CoV-2 mutations associated with severity of COVID-19
- COVID-19 and Breastfeeding
- SARS-CoV-2 vaccine challenge based on spike glycoprotein against several new variants
- Pre-existing Immunity to Endemic Human Coronaviruses Does Not Affect the Immune Response to SARS-CoV-2 Spike in a Murine Vaccination Model
- SARS-CoV-2-Specific T Cell Responses in Patients with COVID-19 and Unexposed Individuals