Clonal Distribution of Clindamycin-Resistant Erythromycin-Susceptible (CRES) Streptococcus agalactiae in Korea Based on Whole Genome Sequences
- Affiliations
-
- 1Laboratory of Infectious Diseases, Graduate School of Infection Control Sciences & Kitasato Institute for Life Sciences, Kitasato University, Tokyo, Japan
- 2Department of Laboratory Medicine, Gyeongsang National University Changwon Hospital, Changwon, Korea
- 3Department of Laboratory Medicine, Gyeongsang National University Hospital, Jinju, Korea
- 4Department of Laboratory Medicine, Gyeongsang National University College of Medicine, Institute of Health Sciences, Jinju, Korea
- KMID: 2502567
- DOI: http://doi.org/10.3343/alm.2020.40.5.370
Abstract
- Background
The clindamycin-resistant erythromycin-susceptible (CRES) phenotype is rare in Streptococcus agalactiae (group B streptococci). We aimed to determine the molecular characteristics of CRES S. agalactiae using whole genome sequencing (WGS).
Methods
Sixty-six S. agalactiae isolates obtained from blood (N=26), cerebrospinal fluid (N=10), urine (N=17), and vaginal discharge (N=13) between 2010 and 2017 in Korea were subjected to WGS. Based on the WGS data, we analyzed antimicrobial resistance (AMR) determinants, sequence types (STs), capsular polysaccharide (CPS) genotypes, and virulence gene profiles, and constructed a phylogenetic tree. We included the clindamycin-susceptible erythromycin-resistant (CSER) phenotype for comparison.
Results
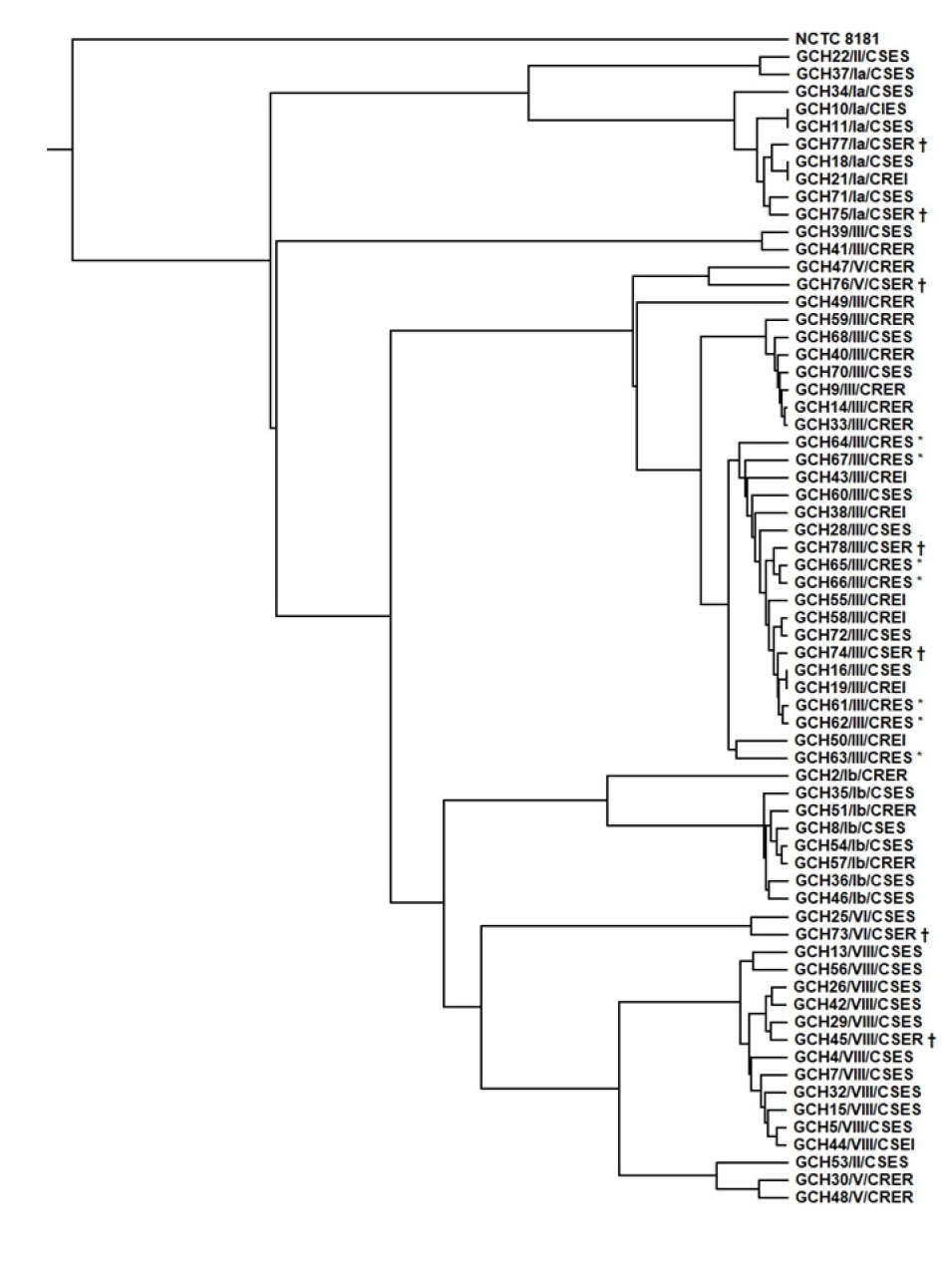
We identified seven CRES S. agalactiae isolates from urine (N=5) and vaginal discharge (N=2) collected between 2010 and 2011. All CRES isolates harbored AMR determinants of lnu(B), lsa(E), and aac(6’)-aph(2’’), revealed ST19 and CPS genotype III, and had a virulence gene profile of rib-lmb-cylE. Phylogenetic tree analysis revealed that all CRES isolates belonged to the same cluster, suggesting a clonal distribution. In contrast, seven CSER isolates showed a diverse distribution and clustered separately from the CRES isolates.
Conclusions
CRES isolates collected between 2010 and 2011 showed a unique cluster with ST19 and CPS genotype III in Korea. This is the first report on WGS-based characteristics of S. agalactiae in Korea.
Keyword
Figure
Cited by 3 articles
-
Comparing Genomic Characteristics of
Streptococcus pyogenes Associated with Invasiveness over a 20-year Period in Korea
Hyoshim Shin, Takashi Takahashi, Seungjun Lee, Eun Hwa Choi, Takahiro Maeda, Yasuto Fukushima, Sunjoo Kim
Ann Lab Med. 2022;42(4):438-446. doi: 10.3343/alm.2022.42.4.438.Performance of BD MAX Group B Streptococcus (GBS) Assay without Enrichment for the Detection of GBS
Sewhan Um, Jaeyoung Her, Si Hyun Kim, Sae Am Song, Young Nam Kim, Jeong Hwan Shin
Ann Lab Med. 2022;42(4):478-481. doi: 10.3343/alm.2022.42.4.478.Virulence-associated Genome Sequences of
Pasteurella canis and Unique Toxin Gene Prevalence ofP. canis andPasteurella multocida Isolated from Humans and Companion Animals
Haruno Yoshida, Jung-Min Kim, Takahiro Maeda, Mieko Goto, Yuzo Tsuyuki, Sachiko Shibata, Kenichi Shizuno, Katsuko Okuzumi, Jae-Seok Kim, Takashi Takahashi
Ann Lab Med. 2023;43(3):263-272. doi: 10.3343/alm.2023.43.3.263.
Reference
-
1. Takahashi T, Sunaoshi K, Sunakawa K, Fujishima S, Watanabe H, Ubukata K, et al. Clinical aspects of invasive infections with Streptococcus dysgalactiae ssp. equisimilis in Japan: differences with respect to Streptococcus pyogenes and Streptococcus agalactiae infections. Clin Microbiol Infect. 2010; 16:1097–103.2. Shibayama A, Yoshizaki T, Tamaki M, Goto M, Takahashi T. Pyogenic sternoclavicular arthritis caused by Streptococcus agalactiae in an elderly adult with diabetes mellitus. J Am Geriatr Soc. 2016; 64:1376–7.3. Choi WH, Park HW, Kim S. Persistence of Group B streptococcus in the urogenital area. Ann Lab Med. 2017; 37:454–6.
Article4. Emaneini M, Mirsalehian A, Beigvierdi R, Fooladi AA, Asadi F, Jabalameli F, et al. High incidence of macrolide and tetracycline resistance among Streptococcus agalactiae strains isolated from clinical samples in Tehran, Iran. Maedica (Buchar). 2014; 9:157–61.5. Lindahl G, Stålhammar-Carlemalm M, Areschoug T. Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin Microbiol Rev. 2005; 18:102–27.6. Oviedo P, Pegels E, Laczeski M, Quiroga M, Vergara M. Phenotypic and genotypic characterization of Streptococcus agalactiae in pregnant women. First study in a province of Argentina. Braz J Microbiol. 2013; 44:253–8.7. Kim S, Lee S, Park H, Kim S. Predominance of emm4 and antibiotic resistance of Streptococcus pyogenes in acute pharyngitis in a southern region of Korea. J Med Microbiol. 2019; 68:1053–8.8. Haenni M, Lupo A, Madec JY. Antimicrobial resistance in Streptococcus spp. Microbiol Spectr. 2018; 6:ARBA-0008-2017.9. Moroi H, Kimura K, Ido A, Banno H, Jin W, Wachino JI, et al. Erythromycin-susceptible but clindamycin-resistant phenotype of clinical ermB-PCR-positive Group B streptococci isolates with IS1216E-inserted ermB. Jpn J Infect Dis. 2019; 72:420–2.10. Seo YS, Srinivasan U, Oh KY, Shin JH, Chae JD, Kim MY, et al. Changing molecular epidemiology of group B streptococcus in Korea. J Korean Med Sci. 2010; 25:817–23.
Article11. Hawkins PA, Law CS, Metcalf BJ, Chochua S, Jackson DM, Westblade LF, et al. Cross-resistance to lincosamides, streptogramins A and pleuromutilins in Streptococcus agalactiae isolates from the USA. J Antimicrob Chemother. 2017; 72:1886–92.12. Yoshida H, Katayama Y, Fukushima Y, Taniyama D, Murata Y, Mizutani T, et al. Draft genome sequence of Streptococcus canis clinical strain TA4, harboring the M-like protein gene and isolated in Japan from a patient with bacteremia. Genome Announc. 2018; 6:e01469–17.
Article13. Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010; 11:119.
Article14. Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012; 67:2640–4.
Article15. Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012; 50:1355–61.
Article16. Nascimento M, Sousa A, Ramirez M, Francisco AP, Carriço JA, Vaz C. PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics. 2017; 33:128–9.
Article17. Murayama SY, Seki C, Sakata H, Sunaoshi K, Nakayama E, Iwata S, et al. Capsular type and antibiotic resistance in Streptococcus agalactiae isolates from patients, ranging from newborns to the elderly, with invasive infections. Antimicrob Agents Chemother. 2009; 53:2650–3.18. Manning SD, Ki M, Marrs CF, Kugeler KJ, Borchardt SM, Baker CJ, et al. The frequency of genes encoding three putative group B streptococcal virulence factors among invasive and colonizing isolates. BMC Infect Dis. 2006; 6:116.
Article19. Spellerberg B, Rozdzinski E, Martin S, Weber-Heynemann J, Schnitzler N, Lütticken R, et al. Lmb, a protein with similarities to the LraI adhesin family, mediates attachment of Streptococcus agalactiae to human laminin. Infect Immun. 1999; 67:871–8.20. Bergseng H, Bevanger L, Rygg M, Bergh K. Real-time PCR targeting the sip gene for detection of group B Streptococcus colonization in pregnant women at delivery. J Med Microbiol. 2007; 56:223–8.
Article21. Lee I, Ouk Kim Y, Park SC, Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016; 66:1100–3.
Article22. PubMLST. Isolate information: id 4644 (593049) - Streptococcus agalactiae isolates. https://pubmlst.org/bigsdb?page=info&db=pubmlst_sagalactiae_isolates&id=4644. (Updated on Oct 2017).23. PubMLST. Isolate information: id 5773 (iGBS308) - Streptococcus agalactiae isolates. https://pubmlst.org/bigsdb?page=info&db=pubmlst_sagalactiae_isolates&id=5773. (Updated on Oct 2019).24. Ryu H, Park YJ, Kim YK, Chang J, Yu JK. Dominance of clonal complex 10 among the levofloxacin-resistant Streptococcus agalactiae isolated from bacteremic patients in a Korean hospital. J Infect Chemother. 2014; 20:509–11.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Erythromycin Resistance Phenotype of Streptococcus pyogenes
- Changing Molecular Epidemiology of Group B Streptococcus in Korea
- Prevalence of Inducible Macrolide-Lincosamide-Streptogramin B (MLS(B)) Resistance in Erythromycin-Resistant Staphylococci
- Perineal Colonization Rate and Antimicrobial Susceptibility of Group B Streptococcus in Pregnant and Non-Pregnant Korean Women
- Macrolide Resistance Trends in beta-Hemolytic Streptococci in a Tertiary Korean Hospital