Endocrinol Metab.
2020 Mar;35(1):44-54. 10.3803/EnM.2020.35.1.44.
Recent Improvements in Genomic and Transcriptomic Understanding of Anaplastic and Poorly Differentiated Thyroid Cancers
- Affiliations
-
- 1Human Oncology and Pathogenesis Program, Memorial Sloan Kettering Cancer Center, New York, NY, USA.
- 2Department of Internal Medicine, CHA Bundang Medical Center, CHA University, Seongnam, Korea.
- 3Department of Internal Medicine, Seoul National University College of Medicine, Seoul, Korea. yjparkmd@snu.ac.kr
- 4Genomic Medicine Institute, Medical Research Center, Seoul National University, Seoul, Korea.
- 5Precision Medicine Center, Seoul National University Bundang Hospital, Seongnam, Korea. jeongsunseo@gmail.com
- 6Gong-Wu Genomic Medicine Institute, Seoul National University Bundang Hospital, Seongnam, Korea.
- 7Macrogen Inc., Seoul, Korea.
- KMID: 2471719
- DOI: http://doi.org/10.3803/EnM.2020.35.1.44
Abstract
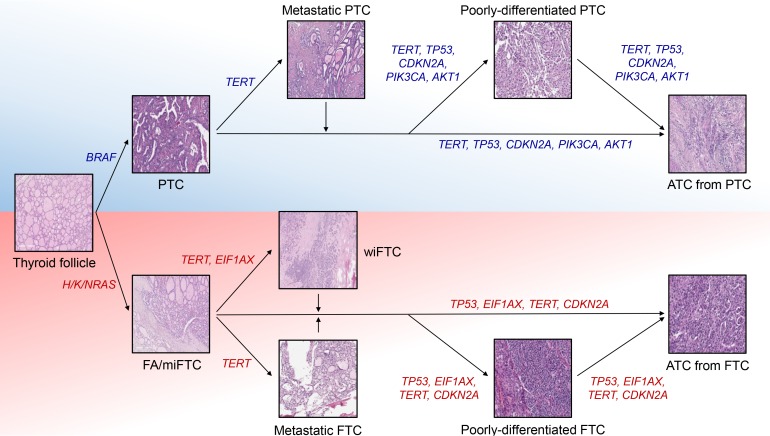
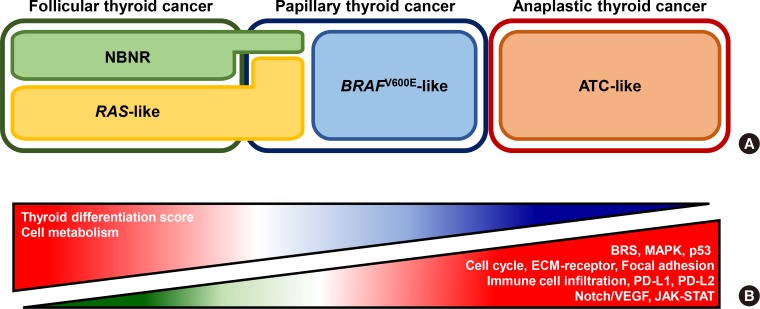
- Anaplastic thyroid cancer (ATC) is a lethal human cancer with a 5-year survival rate of less than 10%. Recently, its genomic and transcriptomic characteristics have been extensively elucidated over 5 years owing to advance in high throughput sequencing. These efforts have extended molecular understandings into the progression mechanisms and therapeutic vulnerabilities of aggressive thyroid cancers. In this review, we provide an overview of genomic and transcriptomic alterations in ATC and poorly-differentiated thyroid cancer, which are distinguished from differentiated thyroid cancers. Clinically relevant genomic alterations and deregulated signaling pathways will be able to shed light on more effective prevention and stratified therapeutic interventions for affected patients.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Mechanisms of
TERT Reactivation and Its Interaction withBRAF V600E
Young Shin Song, Young Joo Park
Endocrinol Metab. 2020;35(3):515-525. doi: 10.3803/EnM.2020.304.
Reference
-
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019; 69:7–34. PMID: 30620402.
Article2. Dralle H, Machens A, Basa J, Fatourechi V, Franceschi S, Hay ID, et al. Follicular cell-derived thyroid cancer. Nat Rev Dis Primers. 2015; 1:15077. PMID: 27188261.
Article3. La Vecchia C, Malvezzi M, Bosetti C, Garavello W, Bertuccio P, Levi F, et al. Thyroid cancer mortality and incidence: a global overview. Int J Cancer. 2015; 136:2187–2195. PMID: 25284703.
Article4. Ibrahimpasic T, Ghossein R, Shah JP, Ganly I. Poorly differentiated carcinoma of the thyroid gland: current status and future prospects. Thyroid. 2019; 29:311–321. PMID: 30747050.
Article5. Hyman DM, Taylor BS, Baselga J. Implementing genome-driven oncology. Cell. 2017; 168:584–599. PMID: 28187282.
Article6. Song YS, Park YJ. Genomic characterization of differentiated thyroid carcinoma. Endocrinol Metab (Seoul). 2019; 34:1–10. PMID: 30912334.
Article7. Bollag G, Tsai J, Zhang J, Zhang C, Ibrahim P, Nolop K, et al. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nat Rev Drug Discov. 2012; 11:873–886. PMID: 23060265.
Article8. Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010; 363:1693–1703. PMID: 20979469.9. Shaw AT, Friboulet L, Leshchiner I, Gainor JF, Bergqvist S, Brooun A, et al. Resensitization to crizotinib by the lorlatinib ALK resistance mutation L1198F. N Engl J Med. 2016; 374:54–61. PMID: 26698910.10. Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014; 159:676–690. PMID: 25417114.11. Yoo SK, Lee S, Kim SJ, Jee HG, Kim BA, Cho H, et al. Comprehensive analysis of the transcriptional and mutational landscape of follicular and papillary thyroid cancers. PLoS Genet. 2016; 12:e1006239. PMID: 27494611.
Article12. Mercer KE, Pritchard CA. Raf proteins and cancer: B-Raf is identified as a mutational target. Biochim Biophys Acta. 2003; 1653:25–40. PMID: 12781369.
Article13. Jung SH, Kim MS, Jung CK, Park HC, Kim SY, Liu J, et al. Mutational burdens and evolutionary ages of thyroid follicular adenoma are comparable to those of follicular carcinoma. Oncotarget. 2016; 7:69638–69648. PMID: 27626165.
Article14. Yoo SK, Song YS, Lee EK, Hwang J, Kim HH, Jung G, et al. Integrative analysis of genomic and transcriptomic characteristics associated with progression of aggressive thyroid cancer. Nat Commun. 2019; 10:2764. PMID: 31235699.
Article15. Duan H, Li Y, Hu P, Gao J, Ying J, Xu W, et al. Mutational profiling of poorly differentiated and anaplastic thyroid carcinoma by the use of targeted next-generation sequencing. Histopathology. 2019; 75:890–899. PMID: 31230400.
Article16. Pozdeyev N, Gay LM, Sokol ES, Hartmaier R, Deaver KE, Davis S, et al. Genetic analysis of 779 advanced differentiated and anaplastic thyroid cancers. Clin Cancer Res. 2018; 24:3059–3068. PMID: 29615459.
Article17. Chen H, Luthra R, Routbort MJ, Patel KP, Cabanillas ME, Broaddus RR, et al. Molecular profile of advanced thyroid carcinomas by next-generation sequencing: characterizing tumors beyond diagnosis for targeted therapy. Mol Cancer Ther. 2018; 17:1575–1584. PMID: 29695638.
Article18. Landa I, Ibrahimpasic T, Boucai L, Sinha R, Knauf JA, Shah RH, et al. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest. 2016; 126:1052–1066. PMID: 26878173.
Article19. Kunstman JW, Juhlin CC, Goh G, Brown TC, Stenman A, Healy JM, et al. Characterization of the mutational landscape of anaplastic thyroid cancer via whole-exome sequencing. Hum Mol Genet. 2015; 24:2318–2329. PMID: 25576899.
Article20. Khan SA, Ci B, Xie Y, Gerber DE, Beg MS, Sherman SI, et al. Unique mutation patterns in anaplastic thyroid cancer identified by comprehensive genomic profiling. Head Neck. 2019; 41:1928–1934. PMID: 30758123.
Article21. Tiedje V, Ting S, Herold T, Synoracki S, Latteyer S, Moeller LC, et al. NGS based identification of mutational hotspots for targeted therapy in anaplastic thyroid carcinoma. Oncotarget. 2017; 8:42613–42620. PMID: 28489587.
Article22. Bonhomme B, Godbert Y, Perot G, Al Ghuzlan A, Bardet S, Belleannee G, et al. Molecular pathology of anaplastic thyroid carcinomas: a retrospective study of 144 cases. Thyroid. 2017; 27:682–692. PMID: 28351340.
Article23. Jeon MJ, Chun SM, Kim D, Kwon H, Jang EK, Kim TY, et al. Genomic alterations of anaplastic thyroid carcinoma detected by targeted massive parallel sequencing in a BRAF (V600E) mutation-prevalent area. Thyroid. 2016; 26:683–690. PMID: 26980298.24. Kelly LM, Barila G, Liu P, Evdokimova VN, Trivedi S, Panebianco F, et al. Identification of the transforming STRN-ALK fusion as a potential therapeutic target in the aggressive forms of thyroid cancer. Proc Natl Acad Sci U S A. 2014; 111:4233–4238. PMID: 24613930.
Article25. Panebianco F, Nikitski AV, Nikiforova MN, Kaya C, Yip L, Condello V, et al. Characterization of thyroid cancer driven by known and novel ALK fusions. Endocr Relat Cancer. 2019; 26:803–814. PMID: 31539879.
Article26. Nikitski AV, Rominski SL, Wankhede M, Kelly LM, Panebianco F, Barila G, et al. Mouse model of poorly differentiated thyroid carcinoma driven by STRN-ALK fusion. Am J Pathol. 2018; 188:2653–2661. PMID: 30125543.
Article27. Landa I, Pozdeyev N, Korch C, Marlow LA, Smallridge RC, Copland JA, et al. Comprehensive genetic characterization of human thyroid cancer cell lines: a validated panel for preclinical studies. Clin Cancer Res. 2019; 25:3141–3151. PMID: 30737244.
Article28. Martin M, Masshofer L, Temming P, Rahmann S, Metz C, Bornfeld N, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013; 45:933–936. PMID: 23793026.
Article29. Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2017; 32:204–220. PMID: 28810145.
Article30. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2:401–404. PMID: 22588877.31. Yu C, Luo C, Qu B, Khudhair N, Gu X, Zang Y, et al. Molecular network including eIF1AX, RPS7, and 14-3-3γ regulates protein translation and cell proliferation in bovine mammary epithelial cells. Arch Biochem Biophys. 2014; 564:142–155. PMID: 25281768.
Article32. Krishnamoorthy GP, Davidson NR, Leach SD, Zhao Z, Lowe SW, Lee G, et al. EIF1AX and RAS mutations cooperate to drive thyroid tumorigenesis through ATF4 and c-MYC. Cancer Discov. 2019; 9:264–281. PMID: 30305285.
Article33. Dienstmann R, Rodon J, Serra V, Tabernero J. Picking the point of inhibition: a comparative review of PI3K/AKT/mTOR pathway inhibitors. Mol Cancer Ther. 2014; 13:1021–1031. PMID: 24748656.
Article34. Millis SZ, Ikeda S, Reddy S, Gatalica Z, Kurzrock R. Landscape of phosphatidylinositol-3-kinase pathway alterations across 19 784 diverse solid tumors. JAMA Oncol. 2016; 2:1565–1573. PMID: 27388585.35. Chaft JE, Arcila ME, Paik PK, Lau C, Riely GJ, Pietanza MC, et al. Coexistence of PIK3CA and other oncogene mutations in lung adenocarcinoma-rationale for comprehensive mutation profiling. Mol Cancer Ther. 2012; 11:485–491. PMID: 22135231.
Article36. Janku F, Lee JJ, Tsimberidou AM, Hong DS, Naing A, Falchook GS, et al. PIK3CA mutations frequently coexist with RAS and BRAF mutations in patients with advanced cancers. PLoS One. 2011; 6:e22769. PMID: 21829508.
Article37. Chiu JW, Krzyzanowska MK, Serra S, Knox JJ, Dhani NC, Mackay H, et al. Molecular profiling of patients with advanced colorectal cancer: princess margaret cancer centre experience. Clin Colorectal Cancer. 2018; 17:73–79. PMID: 29128266.
Article38. Gibson WJ, Ruan DT, Paulson VA, Barletta JA, Hanna GJ, Kraft S, et al. Genomic heterogeneity and exceptional response to dual pathway inhibition in anaplastic thyroid cancer. Clin Cancer Res. 2017; 23:2367–2373. PMID: 27797976.
Article39. Rudolph M, Anzeneder T, Schulz A, Beckmann G, Byrne AT, Jeffers M, et al. AKT1 (E17K) mutation profiling in breast cancer: prevalence, concurrent oncogenic alterations, and blood-based detection. BMC Cancer. 2016; 16:622. PMID: 27515171.
Article40. Bleeker FE, Felicioni L, Buttitta F, Lamba S, Cardone L, Rodolfo M, et al. AKT1(E17K) in human solid tumours. Oncogene. 2008; 27:5648–5650. PMID: 18504432.
Article41. Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007; 448:439–444. PMID: 17611497.42. Porta C, Paglino C, Mosca A. Targeting PI3K/Akt/mTOR signaling in cancer. Front Oncol. 2014; 4:64. PMID: 24782981.
Article44. Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017; 23:703–713. PMID: 28481359.45. Xing M, Liu R, Liu X, Murugan AK, Zhu G, Zeiger MA, et al. BRAF V600E and TERT promoter mutations cooperatively identify the most aggressive papillary thyroid cancer with highest recurrence. J Clin Oncol. 2014; 32:2718–2726. PMID: 25024077.
Article46. Melo M, da Rocha AG, Vinagre J, Batista R, Peixoto J, Tavares C, et al. TERT promoter mutations are a major indicator of poor outcome in differentiated thyroid carcinomas. J Clin Endocrinol Metab. 2014; 99:E754–E765. PMID: 24476079.
Article47. Song YS, Lim JA, Choi H, Won JK, Moon JH, Cho SW, et al. Prognostic effects of TERT promoter mutations are enhanced by coexistence with BRAF or RAS mutations and strengthen the risk prediction by the ATA or TNM staging system in differentiated thyroid cancer patients. Cancer. 2016; 122:1370–1379. PMID: 26969876.48. Liu R, Zhang T, Zhu G, Xing M. Regulation of mutant TERT by BRAF V600E/MAP kinase pathway through FOS/GABP in human cancer. Nat Commun. 2018; 9:579. PMID: 29422527.
Article49. Song YS, Yoo SK, Kim HH, Jung G, Oh AR, Cha JY, et al. Interaction of BRAF-induced ETS factors with mutant TERT promoter in papillary thyroid cancer. Endocr Relat Cancer. 2019; 26:629–641. PMID: 30999281.
Article50. Diplas BH, He X, Brosnan-Cashman JA, Liu H, Chen LH, Wang Z, et al. The genomic landscape of TERT promoter wildtype-IDH wildtype glioblastoma. Nat Commun. 2018; 9:2087. PMID: 29802247.
Article51. Valentijn LJ, Koster J, Zwijnenburg DA, Hasselt NE, van Sluis P, Volckmann R, et al. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat Genet. 2015; 47:1411–1414. PMID: 26523776.
Article52. Peifer M, Hertwig F, Roels F, Dreidax D, Gartlgruber M, Menon R, et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015; 526:700–704. PMID: 26466568.
Article53. Liang WS, Hendricks W, Kiefer J, Schmidt J, Sekar S, Carpten J, et al. Integrated genomic analyses reveal frequent TERT aberrations in acral melanoma. Genome Res. 2017; 27:524–532. PMID: 28373299.
Article54. Donehower LA, Soussi T, Korkut A, Liu Y, Schultz A, Cardenas M, et al. Integrated analysis of TP53 gene and pathway alterations in the Cancer Genome Atlas. Cell Rep. 2019; 28:1370–1384. PMID: 31365877.
Article55. Cho SY, Park C, Na D, Han JY, Lee J, Park OK, et al. High prevalence of TP53 mutations is associated with poor survival and an EMT signature in gliosarcoma patients. Exp Mol Med. 2017; 49:e317. PMID: 28408749.
Article56. Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014; 25:304–317. PMID: 24651012.
Article57. Acin S, Li Z, Mejia O, Roop DR, El-Naggar AK, Caulin C. Gain-of-function mutant p53 but not p53 deletion promotes head and neck cancer progression in response to oncogenic K-ras. J Pathol. 2011; 225:479–489. PMID: 21952947.
Article58. McFadden DG, Vernon A, Santiago PM, Martinez-McFaline R, Bhutkar A, Crowley DM, et al. p53 constrains progression to anaplastic thyroid carcinoma in a Braf-mutant mouse model of papillary thyroid cancer. Proc Natl Acad Sci U S A. 2014; 111:E1600–E1609. PMID: 24711431.
Article59. Liggett WH Jr, Sidransky D. Role of the p16 tumor suppressor gene in cancer. J Clin Oncol. 1998; 16:1197–1206. PMID: 9508208.
Article60. Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012; 489:519–525. PMID: 22960745.61. Bui NQ, Przybyl J, Trabucco SE, Frampton G, Hastie T, van de Rijn M, et al. A clinico-genomic analysis of soft tissue sarcoma patients reveals CDKN2A deletion as a biomarker for poor prognosis. Clin Sarcoma Res. 2019; 9:12. PMID: 31528332.
Article62. Shain AH, Yeh I, Kovalyshyn I, Sriharan A, Talevich E, Gagnon A, et al. The genetic evolution of melanoma from precursor lesions. N Engl J Med. 2015; 373:1926–1936. PMID: 26559571.
Article63. Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015; 160:1246–1260. PMID: 25748654.
Article64. Ravi N, Yang M, Gretarsson S, Jansson C, Mylona N, Sydow SR, et al. Identification of targetable lesions in anaplastic thyroid cancer by genome profiling. Cancers (Basel). 2019; 11:E402. PMID: 30909364.
Article65. Paulsson JO, Backman S, Wang N, Stenman A, Crona J, Thutkawkorapin J, et al. Whole-genome sequencing of synchronous thyroid carcinomas identifies aberrant DNA repair in thyroid cancer dedifferentiation. J Pathol. 2020; 250:183–194. PMID: 31621921.
Article66. Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017; 357:409–413. PMID: 28596308.67. Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology: mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015; 348:124–128. PMID: 25765070.68. Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015; 372:2509–2520. PMID: 26028255.69. Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014; 371:2189–2199. PMID: 25409260.
Article70. Wang S, Jia M, He Z, Liu XS. APOBEC3B and APOBEC mutational signature as potential predictive markers for immunotherapy response in non-small cell lung cancer. Oncogene. 2018; 37:3924–3936. PMID: 29695832.
Article71. Faden DL, Ding F, Lin Y, Zhai S, Kuo F, Chan TA, et al. APOBEC mutagenesis is tightly linked to the immune landscape and immunotherapy biomarkers in head and neck squamous cell carcinoma. Oral Oncol. 2019; 96:140–147. PMID: 31422205.
Article72. Kasaian K, Wiseman SM, Walker BA, Schein JE, Zhao Y, Hirst M, et al. The genomic and transcriptomic landscape of anaplastic thyroid cancer: implications for therapy. BMC Cancer. 2015; 15:984. PMID: 26680454.
Article73. Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013; 14:1014–1022. PMID: 24048123.
Article74. Kim K, Jeon S, Kim TM, Jung CK. Immune gene signature delineates a subclass of papillary thyroid cancer with unfavorable clinical outcomes. Cancers (Basel). 2018; 10:E494. PMID: 30563160.
Article75. Coates PJ, Rundle JK, Lorimore SA, Wright EG. Indirect macrophage responses to ionizing radiation: implications for genotype-dependent bystander signaling. Cancer Res. 2008; 68:450–456. PMID: 18199539.
Article76. Giannini R, Moretti S, Ugolini C, Macerola E, Menicali E, Nucci N, et al. Immune profiling of thyroid carcinomas suggests the existence of two major phenotypes: an ATC-like and a PDTC-like. J Clin Endocrinol Metab. 2019; 104:3557–3575. PMID: 30882858.
Article77. Galon J, Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat Rev Drug Discov. 2019; 18:197–218. PMID: 30610226.
Article78. Cantara S, Bertelli E, Occhini R, Regoli M, Brilli L, Pacini F, et al. Blockade of the programmed death ligand 1 (PD-L1) as potential therapy for anaplastic thyroid cancer. Endocrine. 2019; 64:122–129. PMID: 30762153.
Article79. Chintakuntlawar AV, Rumilla KM, Smith CY, Jenkins SM, Foote RL, Kasperbauer JL, et al. Expression of PD-1 and PD-L1 in anaplastic thyroid cancer patients treated with multimodal therapy: results from a retrospective study. J Clin Endocrinol Metab. 2017; 102:1943–1950. PMID: 28324060.
Article80. Ahn S, Kim TH, Kim SW, Ki CS, Jang HW, Kim JS, et al. Comprehensive screening for PD-L1 expression in thyroid cancer. Endocr Relat Cancer. 2017; 24:97–106. PMID: 28093480.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Recent Progress of Genome Study for Anaplastic Thyroid Cancer
- Redifferentiation Therapy in Thyroid Cancer
- What's New in Molecular Targeted Therapies for Thyroid Cancer?
- Genomic Characterization of Differentiated Thyroid Carcinoma
- Anaplastic Transformation of Papillary Thyroid Carcinoma in a Young Man: A Case Study with Immunohistochemical and BRAF Analysis