Int J Stem Cells.
2019 Jul;12(2):340-346. 10.15283/ijsc18123.
Hybrid Nanofiber Scaffold-Based Direct Conversion of Neural Precursor Cells/Dopamine Neurons
- Affiliations
-
- 1Research and Development Center, Jeil Pharmaceutical Company, Yongin, Korea.
- 2Graduate School of Biomedical Science & Engineering, Hanyang University, Seoul, Korea. chshpark@hanyang.ac.kr
- 3School of Mechanical & Aerospace Engineering, Seoul National University, Seoul, Korea.
- 4Center for Food and Bioconvergence, Department of Food Science and Biotechnology, Seoul National University, Seoul, Korea.
- 5Research Institute of Advanced Materials, Seoul National University, Seoul, Korea. keesung@snu.ac.kr
- 6Department of Microbiology, College of Medicine, Hanyang University, Seoul, Korea.
- KMID: 2465903
- DOI: http://doi.org/10.15283/ijsc18123
Abstract
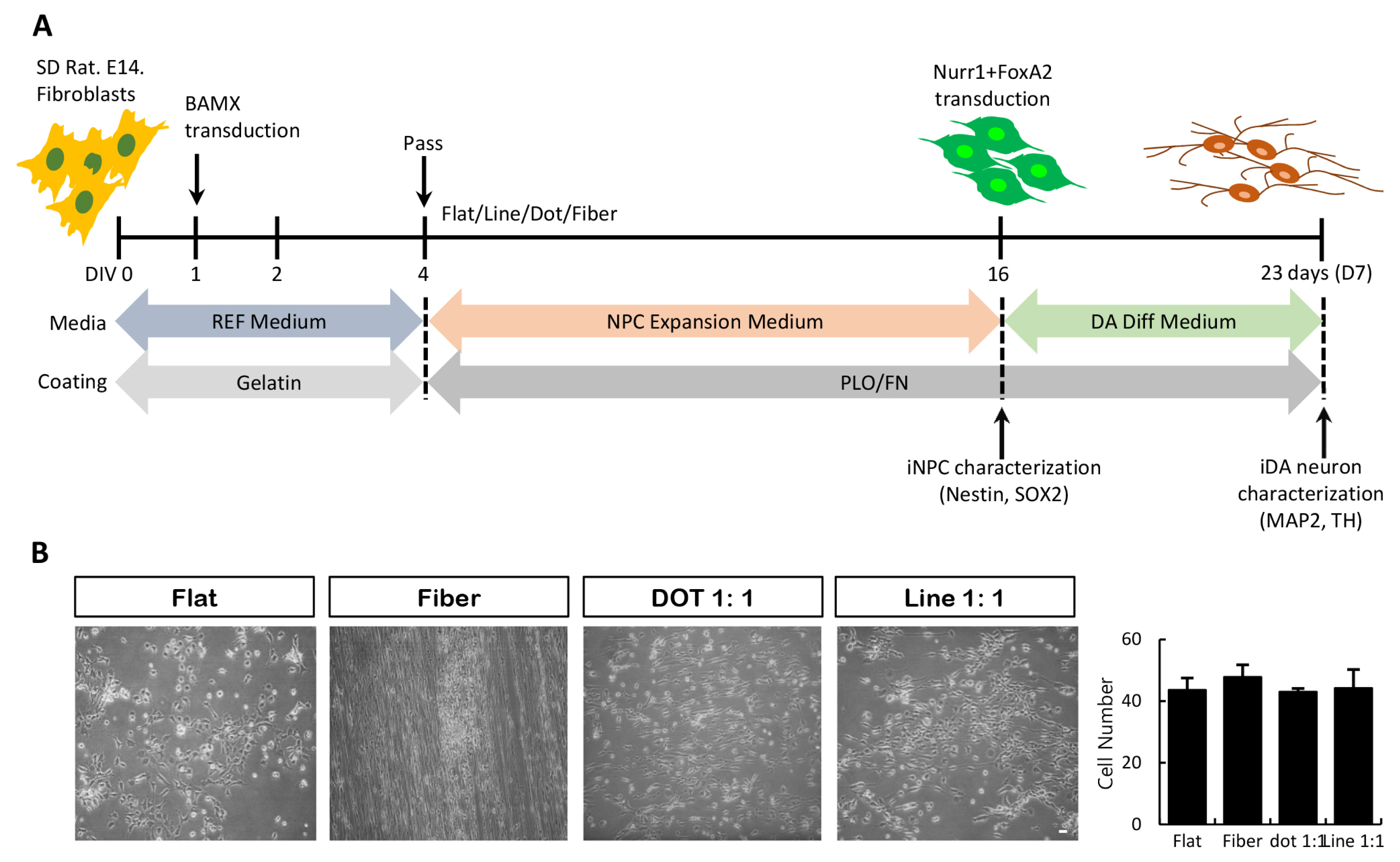
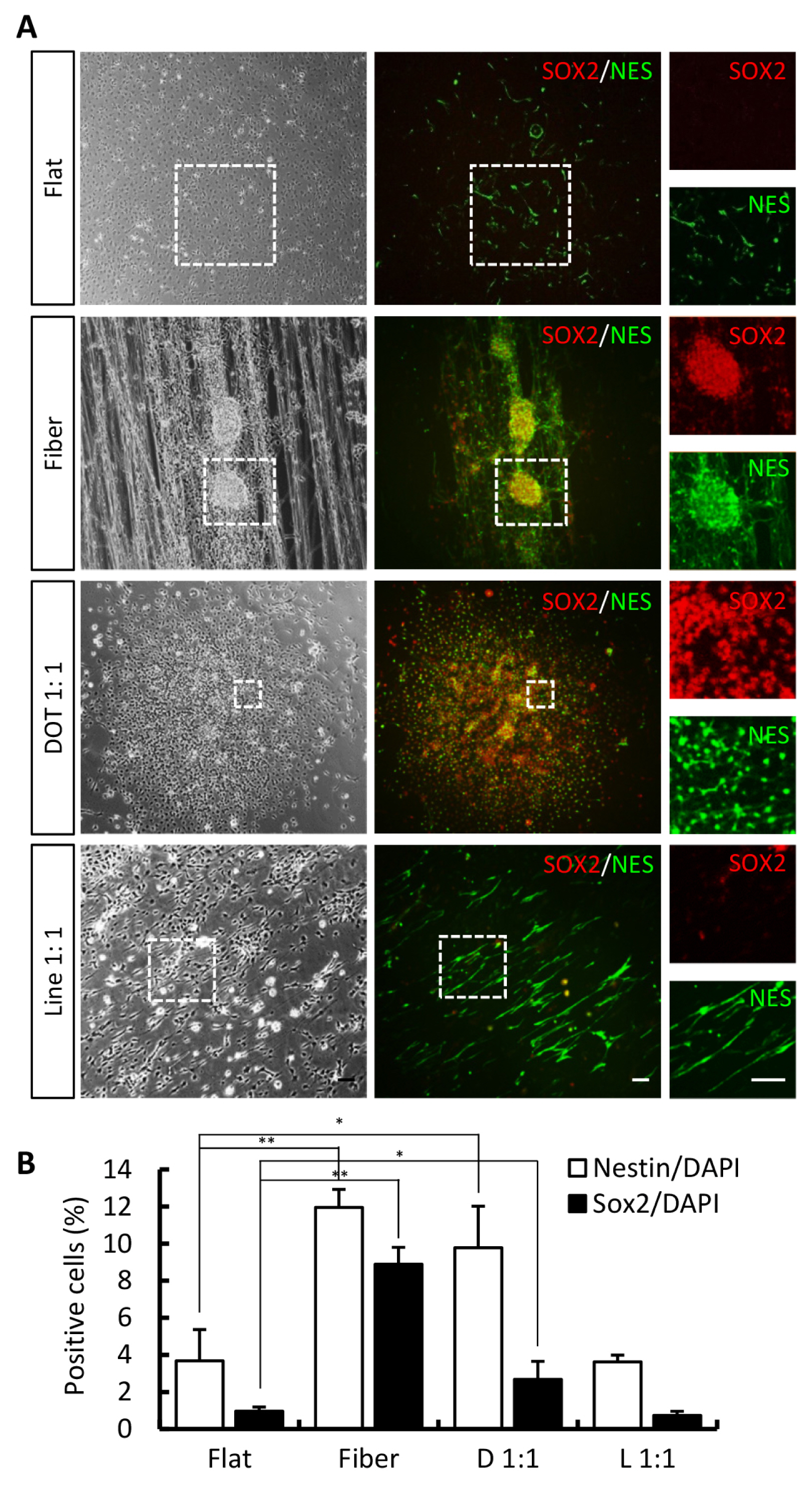
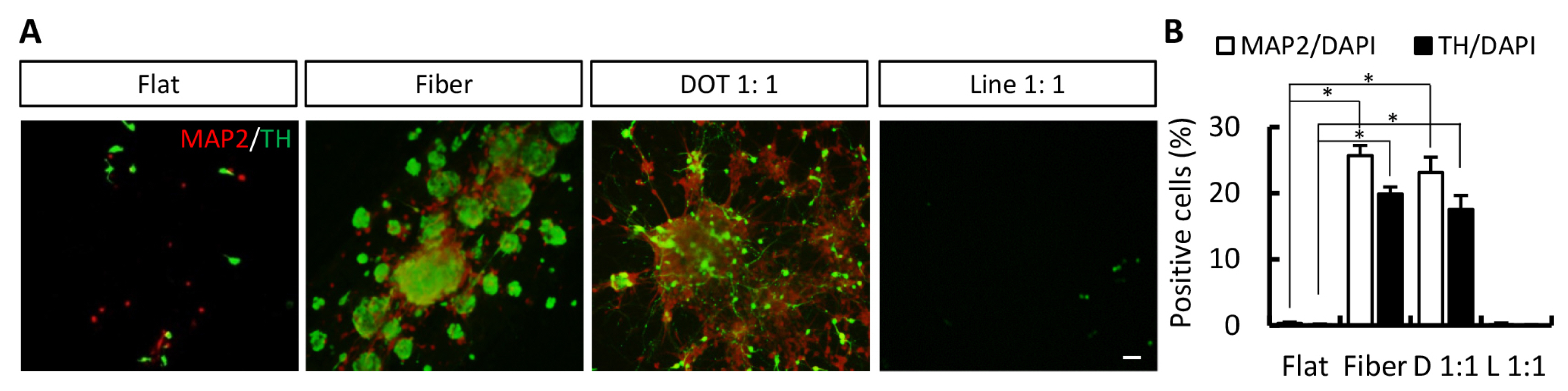
- The concept of cellular reprogramming was developed to generate induced neural precursor cells (iNPCs)/dopaminergic (iDA) neurons using diverse approaches. Here, we investigated the effects of various nanoscale scaffolds (fiber, dot, and line) on iNPC/iDA differentiation by direct reprogramming. The generation and maturation of iDA neurons (microtubule-associated protein 2-positive and tyrosine hydroxylase-positive) and iNPCs (NESTIN-positive and SOX2-positive) increased on fiber and dot scaffolds as compared to that of the flat (control) scaffold. This study demonstrates that nanotopographical environments are suitable for direct differentiation methods and may improve the differentiation efficiency.
Keyword
Figure
Reference
-
References
1. Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010; 463:1035–1041. DOI: 10.1038/nature08797. PMID: 20107439. PMCID: PMC2829121.
Article2. Kim J, Efe JA, Zhu S, Talantova M, Yuan X, Wang S, Lipton SA, Zhang K, Ding S. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci U S A. 2011; 108:7838–7843. DOI: 10.1073/pnas.1103113108. PMID: 21521790. PMCID: PMC3093517.
Article3. Thier M, Wörsdörfer P, Lakes YB, Gorris R, Herms S, Opitz T, Seiferling D, Quandel T, Hoffmann P, Nöthen MM, Brüstle O, Edenhofer F. Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell. 2012; 10:473–479. DOI: 10.1016/j.stem.2012.03.003. PMID: 22445518.
Article4. Lim MS, Chang MY, Kim SM, Yi SH, Suh-Kim H, Jung SJ, Kim MJ, Kim JH, Lee YS, Lee SY, Kim DW, Lee SH, Park CH. Generation of dopamine neurons from rodent fibroblasts through the expandable neural precursor cell stage. J Biol Chem. 2015; 290:17401–17414. DOI: 10.1074/jbc.M114.629808. PMID: 26023233. PMCID: PMC4498077.
Article5. Lim MS, Lee SY, Park CH. FGF8 is essential for functionality of induced neural precursor cell-derived dopaminergic neurons. Int J Stem Cells. 2015; 8:228–234. DOI: 10.15283/ijsc.2015.8.2.228. PMID: 26634071. PMCID: PMC4651287.
Article6. Kim SM, Kim JW, Kwak TH, Park SW, Kim KP, Park H, Lim KT, Kang K, Kim J, Yang JH, Han H, Lee I, Hyun JK, Bae YM, Schöler HR, Lee HT, Han DW. Generation of integration-free induced neural stem cells from mouse fibroblasts. J Biol Chem. 2016; 291:14199–14212. DOI: 10.1074/jbc.M115.713578. PMID: 27189941. PMCID: PMC4933177.
Article7. Kim J, Kim KP, Lim KT, Lee SC, Yoon J, Song G, Hwang SI, Schöler HR, Cantz T, Han DW. Generation of integration-free induced hepatocyte-like cells from mouse fibroblasts. Sci Rep. 2015; 5:15706. DOI: 10.1038/srep15706. PMID: 26503743. PMCID: PMC4621602.
Article8. Li X, Zuo X, Jing J, Ma Y, Wang J, Liu D, Zhu J, Du X, Xiong L, Du Y, Xu J, Xiao X, Wang J, Chai Z, Zhao Y, Deng H. Small-molecule-driven direct reprogramming of mouse fibroblasts into functional neurons. Cell Stem Cell. 2015; 17:195–203. DOI: 10.1016/j.stem.2015.06.003. PMID: 26253201.
Article9. Zheng J, Choi KA, Kang PJ, Hyeon S, Kwon S, Moon JH, Hwang I, Kim YI, Kim YS, Yoon BS, Park G, Lee J, Hong S, You S. A combination of small molecules directly reprograms mouse fibroblasts into neural stem cells. Biochem Biophys Res Commun. 2016; 476:42–48. DOI: 10.1016/j.bbrc.2016.05.080. PMID: 27207831.
Article10. Lim MS, Kim SM, Lee EH, Park CH. Efficient induction of neural precursor cells from fibroblasts using stromal cell-derived inducing activity. Tissue Eng Regen Med. 2016; 13:554–559. DOI: 10.1007/s13770-016-0012-3. PMID: 30603436. PMCID: PMC6170833.
Article11. Yim EK, Darling EM, Kulangara K, Guilak F, Leong KW. Nanotopography-induced changes in focal adhesions, cytoskeletal organization, and mechanical properties of human mesenchymal stem cells. Biomaterials. 2010; 31:1299–1306. DOI: 10.1016/j.biomaterials.2009.10.037. PMID: 19879643. PMCID: PMC2813896.
Article12. McMurray RJ, Gadegaard N, Tsimbouri PM, Burgess KV, McNamara LE, Tare R, Murawski K, Kingham E, Oreffo RO, Dalby MJ. Nanoscale surfaces for the long-term maintenance of mesenchymal stem cell phenotype and multipotency. Nat Mater. 2011; 10:637–644. DOI: 10.1038/nmat3058. PMID: 21765399.
Article13. Kulangara K, Adler AF, Wang H, Chellappan M, Hammett E, Yasuda R, Leong KW. The effect of substrate topography on direct reprogramming of fibroblasts to induced neurons. Biomaterials. 2014; 35:5327–5336. DOI: 10.1016/j.biomaterials.2014.03.034. PMID: 24709523. PMCID: PMC4023960.
Article14. Yoo J, Noh M, Kim H, Jeon NL, Kim BS, Kim J. Nanogrooved substrate promotes direct lineage reprogramming of fibroblasts to functional induced dopaminergic neurons. Biomaterials. 2015; 45:36–45. DOI: 10.1016/j.biomaterials.2014.12.049. PMID: 25662493.
Article15. Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005; 310:1139–1143. DOI: 10.1126/science.1116995. PMID: 16293750.
Article16. Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006; 126:677–689. DOI: 10.1016/j.cell.2006.06.044. PMID: 16923388.
Article17. Anderson JM, Kushwaha M, Tambralli A, Bellis SL, Camata RP, Jun HW. Osteogenic differentiation of human mesenchymal stem cells directed by extracellular matrix-mimicking ligands in a biomimetic self-assembled peptide amphiphile nanomatrix. Biomacromolecules. 2009; 10:2935–2944. DOI: 10.1021/bm9007452. PMID: 19746964. PMCID: PMC2760643.
Article18. Kurpinski K, Chu J, Hashi C, Li S. Anisotropic mechanosensing by mesenchymal stem cells. Proc Natl Acad Sci U S A. 2006; 103:16095–16100. DOI: 10.1073/pnas.0604182103. PMID: 17060641. PMCID: PMC1637542.
Article19. O’Cearbhaill ED, Punchard MA, Murphy M, Barry FP, McHugh PE, Barron V. Response of mesenchymal stem cells to the biomechanical environment of the endothelium on a flexible tubular silicone substrate. Biomaterials. 2008; 29:1610–1619. DOI: 10.1016/j.biomaterials.2007.11.042. PMID: 18194813.
Article20. Ruiz SA, Chen CS. Emergence of patterned stem cell differentiation within multicellular structures. Stem Cells. 2008; 26:2921–2927. DOI: 10.1634/stemcells.2008-0432. PMID: 18703661. PMCID: PMC2693100.
Article21. McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004; 6:483–495. DOI: 10.1016/S1534-5807(04)00075-9. PMID: 15068789.
Article22. Park J, Cho CH, Parashurama N, Li Y, Berthiaume F, Toner M, Tilles AW, Yarmush ML. Microfabrication-based modulation of embryonic stem cell differentiation. Lab Chip. 2007; 7:1018–1028. DOI: 10.1039/b704739h. PMID: 17653344.
Article23. Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, Wilkinson CD, Oreffo RO. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat Mater. 2007; 6:997–1003. DOI: 10.1038/nmat2013. PMID: 17891143.
Article24. Oh S, Brammer KS, Li YS, Teng D, Engler AJ, Chien S, Jin S. Stem cell fate dictated solely by altered nanotube dimension. Proc Natl Acad Sci U S A. 2009; 106:2130–2135. DOI: 10.1073/pnas.0813200106. PMID: 19179282. PMCID: PMC2650120.
Article25. Lee MR, Kwon KW, Jung H, Kim HN, Suh KY, Kim K, Kim KS. Direct differentiation of human embryonic stem cells into selective neurons on nanoscale ridge/groove pattern arrays. Biomaterials. 2010; 31:4360–4366. DOI: 10.1016/j.biomaterials.2010.02.012. PMID: 20202681.
Article26. Kim HN, Kang DH, Kim MS, Jiao A, Kim DH, Suh KY. Patterning methods for polymers in cell and tissue engineering. Ann Biomed Eng. 2012; 40:1339–1355. DOI: 10.1007/s10439-012-0510-y. PMID: 22258887. PMCID: PMC5439960.
Article27. Li H, Wen F, Chen H, Pal M, Lai Y, Zhao AZ, Tan LP. Micropatterning extracellular matrix proteins on electrospun fibrous substrate promote human mesenchymal stem cell differentiation toward neurogenic lineage. ACS Appl Mater Interfaces. 2016; 8:563–573. DOI: 10.1021/acsami.5b09588. PMID: 26654444.
Article28. You MH, Kwak MK, Kim DH, Kim K, Levchenko A, Kim DY, Suh KY. Synergistically enhanced osteogenic differentiation of human mesenchymal stem cells by culture on nanostructured surfaces with induction media. Biomacromolecules. 2010; 11:1856–1862. DOI: 10.1021/bm100374n. PMID: 20568737. PMCID: PMC2921863.
Article29. Roberts JN, Sahoo JK, McNamara LE, Burgess KV, Yang J, Alakpa EV, Anderson HJ, Hay J, Turner LA, Yarwood SJ, Zelzer M, Oreffo RO, Ulijn RV, Dalby MJ. Dynamic surfaces for the study of mesenchymal stem cell growth through adhesion regulation. ACS Nano. 2016; 10:6667–6679. DOI: 10.1021/acsnano.6b01765. PMID: 27322014. PMCID: PMC4963921.
Article30. Ventre M, Netti PA. Engineering cell instructive materials to control cell fate and functions through material cues and surface patterning. ACS Appl Mater Interfaces. 2016; 8:14896–14908. DOI: 10.1021/acsami.5b08658. PMID: 26693600.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- FGF8 is Essential for Functionality of Induced Neural Precursor Cell-derived Dopaminergic Neurons
- Effect of different sterilization methods on the surface morphology of PPDO-hybrid-PLGA nanofiber scaffold and attachments of PC12 cell
- The Study of the Regeneration of Nucleus Pulposususing Nanofiber-based Scaffold : In Vitro and Animal Study
- Efficient Induction of Neural Precursor Cells from Fibroblasts Using Stromal Cell-Derived Inducing Activity
- Effects of Dopamine on the Gonadotropin Releasing Hormone(GnRH) Neurons