Cancer Res Treat.
2016 Apr;48(2):668-675. 10.4143/crt.2014.288.
Validation of Prediction Models for Mismatch Repair Gene Mutations in Koreans
- Affiliations
-
- 1Department of Surgery, Chonnam National University Hwasun Hospital, Chonnam National University Medical School, Hwasun, Korea.
- 2Department of Surgery, Seoul National University Bundang Hospital, Seongnam, Korea. kdw@snubh.org
- 3Korean Hereditary Tumor Registry, Cancer Research Institute, Seoul National University College of Medicine, Seoul, Korea.
- 4Department of Surgery, Chung Hospital, Seongnam, Korea.
- 5Medical Research Collaborating Center, Seoul National University Bundang Hospital, Seongnam, Korea.
- 6Department of Public Health Science, Seoul National University, Seoul, Korea.
- KMID: 2454345
- DOI: http://doi.org/10.4143/crt.2014.288
Abstract
- PURPOSE
Lynch syndrome, the commonest hereditary colorectal cancer syndrome, is caused by germline mutations in mismatch repair (MMR) genes. Three recently developed prediction models for MMR gene mutations based on family history and clinical features (MMRPredict, PREMM1,2,6, and MMRPro) have been validated only in Western countries. In this study, we propose validating these prediction models in the Korean population.
MATERIALS AND METHODS
We collected MMR gene analysis data from 188 individuals in the Korean Hereditary Tumor Registry. The probability of gene mutation was calculated using three prediction models, and the overall diagnostic value of each model compared using receiver operator characteristic (ROC) curves and area under the ROC curve (AUC). Quantitative test characteristics were calculated at sensitivities of 90%, 95%, and 98%.
RESULTS
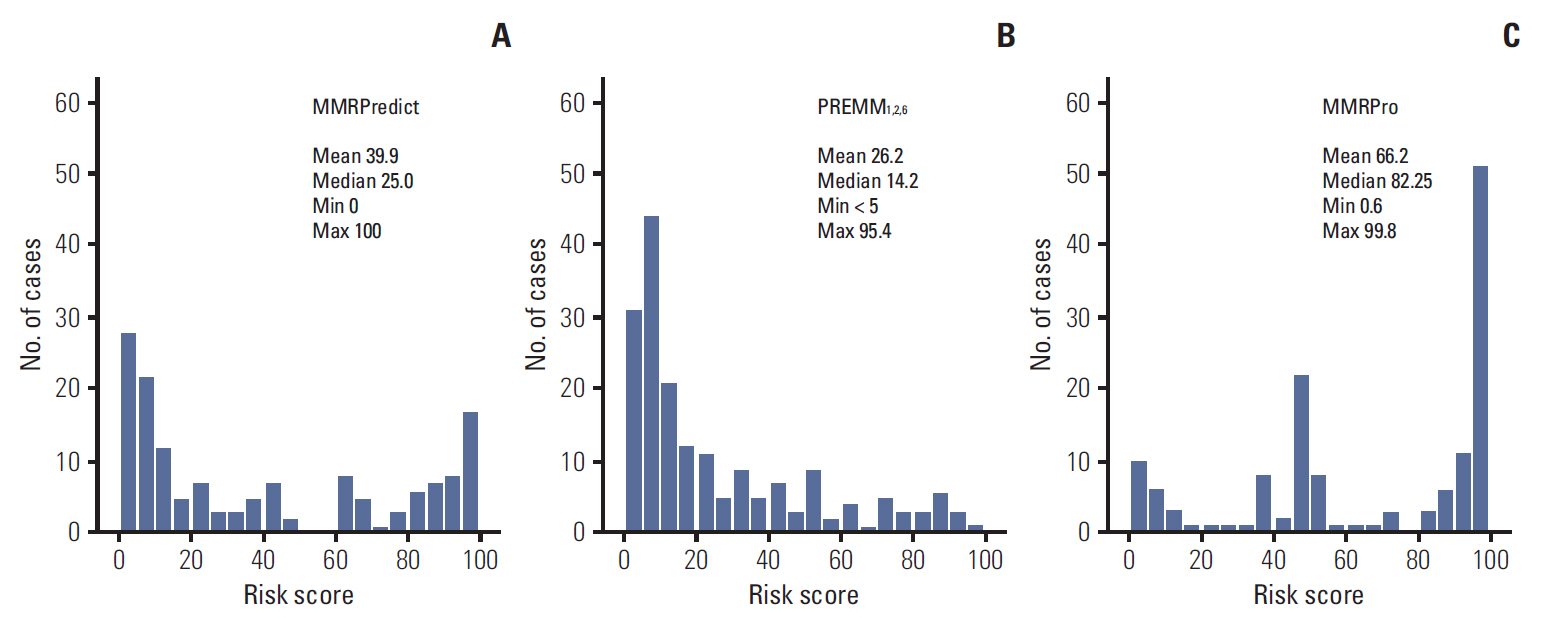
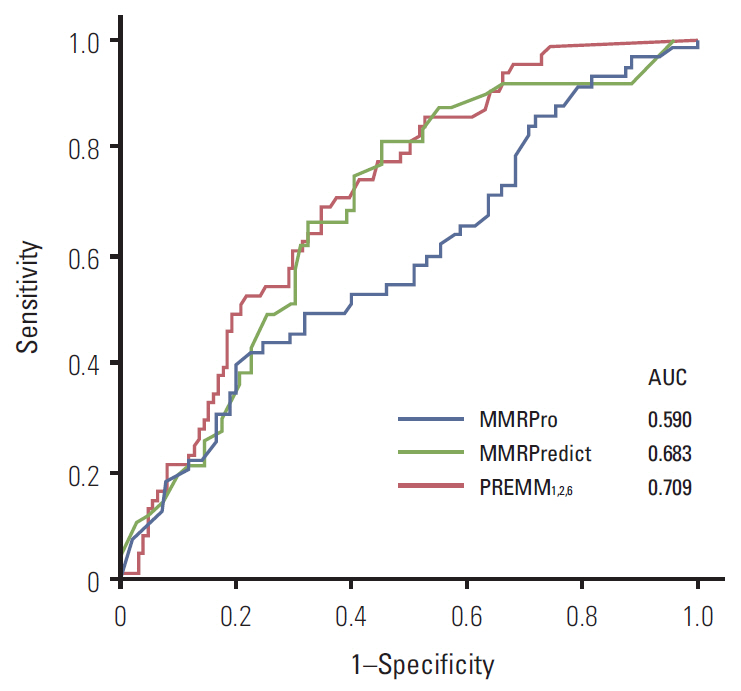
Of the individuals analyzed, 101 satisfied Amsterdam criteria II, and 87 were suspected hereditary nonpolyposis colorectal cancer. MMR mutations were identified in 62 of the 188 subjects (33.0%). All three prediction models showed a poor predictive value of AUC (MMRPredict, 0.683; PREMM1,2,6, 0.709; MMRPro, 0.590). Within the range of acceptable sensitivity (> 90%), PREMM1,2,6 demonstrated higher specificity than the other models.
CONCLUSION
In the Korean population, overall predictive values of the three models (MMRPredict, PREMM1,2,6, MMRPro) for MMR gene mutations are poor, compared with their performance in Western populations. A new prediction model is therefore required for the Korean population to detect MMR mutation carriers, reflecting ethnic differences in genotype-phenotype associations.
Keyword
MeSH Terms
Figure
Reference
-
References
1. Rustgi AK. The genetics of hereditary colon cancer. Genes Dev. 2007; 21:2525–38.
Article2. Lindor NM, Rabe K, Petersen GM, Haile R, Casey G, Baron J, et al. Lower cancer incidence in Amsterdam-I criteria families without mismatch repair deficiency: familial colorectal cancer type X. JAMA. 2005; 293:1979–85.3. Mensenkamp AR, Vogelaar IP, van Zelst-Stams WA, Goossens M, Ouchene H, Hendriks-Cornelissen SJ, et al. Somatic mutations in MLH1 and MSH2 are a frequent cause of mismatch-repair deficiency in Lynch syndrome-like tumors. Gastroenterology. 2014; 146:643–6.
Article4. Bosman FT, Carneiro F, Hruban RH, Theise ND. WHO classification of tumours of the digestive system. 4th ed. Lyon: IARC Press;2010.5. Kastrinos F, Balmana J, Syngal S. Prediction models in Lynch syndrome. Fam Cancer. 2013; 12:217–28.
Article6. Monzon JG, Cremin C, Armstrong L, Nuk J, Young S, Horsman DE, et al. Validation of predictive models for germline mutations in DNA mismatch repair genes in colorectal cancer. Int J Cancer. 2010; 126:930–9.
Article7. Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999; 116:1453–6.
Article8. Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Ruschoff J, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004; 96:261–8.
Article9. Perez-Carbonell L, Ruiz-Ponte C, Guarinos C, Alenda C, Paya A, Brea A, et al. Comparison between universal molecular screening for Lynch syndrome and revised Bethesda guidelines in a large population-based cohort of patients with colorectal cancer. Gut. 2012; 61:865–72.
Article10. Parsons MT, Buchanan DD, Thompson B, Young JP, Spurdle AB. Correlation of tumour BRAF mutations and MLH1 methylation with germline mismatch repair (MMR) gene mutation status: a literature review assessing utility of tumour features for MMR variant classification. J Med Genet. 2012; 49:151–7.11. Pouchet CJ, Wong N, Chong G, Sheehan MJ, Schneider G, Rosen-Sheidley B, et al. A comparison of models used to predict MLH1, MSH2 and MSH6 mutation carriers. Ann Oncol. 2009; 20:681–8.
Article12. Barnetson RA, Tenesa A, Farrington SM, Nicholl ID, Cetnarskyj R, Porteous ME, et al. Identification and survival of carriers of mutations in DNA mismatch-repair genes in colon cancer. N Engl J Med. 2006; 354:2751–63.
Article13. Chen S, Wang W, Lee S, Nafa K, Lee J, Romans K, et al. Prediction of germline mutations and cancer risk in the Lynch syndrome. JAMA. 2006; 296:1479–87.
Article14. Kastrinos F, Steyerberg EW, Mercado R, Balmana J, Holter S, Gallinger S, et al. The PREMM1,2,6 model predicts risk of MLH1, MSH2, and MSH6 germline mutations based on cancer history. Gastroenterology. 2011; 140:73–81.15. Green RC, Parfrey PS, Woods MO, Younghusband HB. Prediction of Lynch syndrome in consecutive patients with colorectal cancer. J Natl Cancer Inst. 2009; 101:331–40.
Article16. Balmana J, Balaguer F, Castellvi-Bel S, Steyerberg EW, Andreu M, Llor X, et al. Comparison of predictive models, clinical criteria and molecular tumour screening for the identification of patients with Lynch syndrome in a population-based cohort of colorectal cancer patients. J Med Genet. 2008; 45:557–63.
Article17. Khan O, Blanco A, Conrad P, Gulden C, Moss TZ, Olopade OI, et al. Performance of Lynch syndrome predictive models in a multi-center US referral population. Am J Gastroenterol. 2011; 106:1822–7.
Article18. Tresallet C, Brouquet A, Julie C, Beauchet A, Vallot C, Menegaux F, et al. Evaluation of predictive models in daily practice for the identification of patients with Lynch syndrome. Int J Cancer. 2012; 130:1367–77.
Article19. Kastrinos F, Steyerberg EW, Balmana J, Mercado R, Gallinger S, Haile R, et al. Comparison of the clinical prediction model PREMM(1,2,6) and molecular testing for the systematic identification of Lynch syndrome in colorectal cancer. Gut. 2013; 62:272–9.20. Shin YK, Heo SC, Shin JH, Hong SH, Ku JL, Yoo BC, et al. Germline mutations in MLH1, MSH2 and MSH6 in Korean hereditary non-polyposis colorectal cancer families. Hum Mutat. 2004; 24:351.21. Oh JR, Kim DW, Lee HS, Lee HE, Lee SM, Jang JH, et al. Microsatellite instability testing in Korean patients with colorectal cancer. Fam Cancer. 2012; 11:459–66.
Article22. De Jesus-Monge WE, Gonzalez-Keelan C, Zhao R, Hamilton SR, Rodriguez-Bigas M, Cruz-Correa M. Mismatch repair protein expression and colorectal cancer in Hispanics from Puerto Rico. Fam Cancer. 2010; 9:155–66.
Article23. Park JG, Vasen HF, Park YJ, Park KJ, Peltomaki P, de Leon MP, et al. Suspected HNPCC and Amsterdam criteria II: evaluation of mutation detection rate, an international collaborative study. Int J Colorectal Dis. 2002; 17:109–14.
Article24. Steyerberg EW, Vickers AJ, Cook NR, Gerds T, Gonen M, Obuchowski N, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology. 2010; 21:128–38.25. Win AK, Macinnis RJ, Dowty JG, Jenkins MA. Criteria and prediction models for mismatch repair gene mutations: a review. J Med Genet. 2013; 50:785–93.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Gastric Cancer and DNA Mismatch Repair
- A Case of Muir-Torre Syndrome: Extra-ocular Sebaceous Carcinoma in a Patient with Breast Cancer
- Expression Pattern of DNA Mismatch Repair Genes in Tumors of Microsatellite Mutator Phenotype
- Hereditary Nonpolyposis Colorectal Cancer
- Hereditary Colon Cancer: Lynch Syndrome