Immune Netw.
2017 Dec;17(6):451-459. 10.4110/in.2017.17.6.451.
ELISA for Quantitative Determination of Hepatitis B Virus Surface Antigen
- Affiliations
-
- 1Division of Biomedical Convergence, College of Biomedical Science, Kangwon National University, Chuncheon 24341, Korea. kimsho@kangwon.ac.kr
- 2Institute of Bioscience and Biotechnology, Kangwon National University, Chuncheon 24341, Korea.
- KMID: 2400630
- DOI: http://doi.org/10.4110/in.2017.17.6.451
Abstract
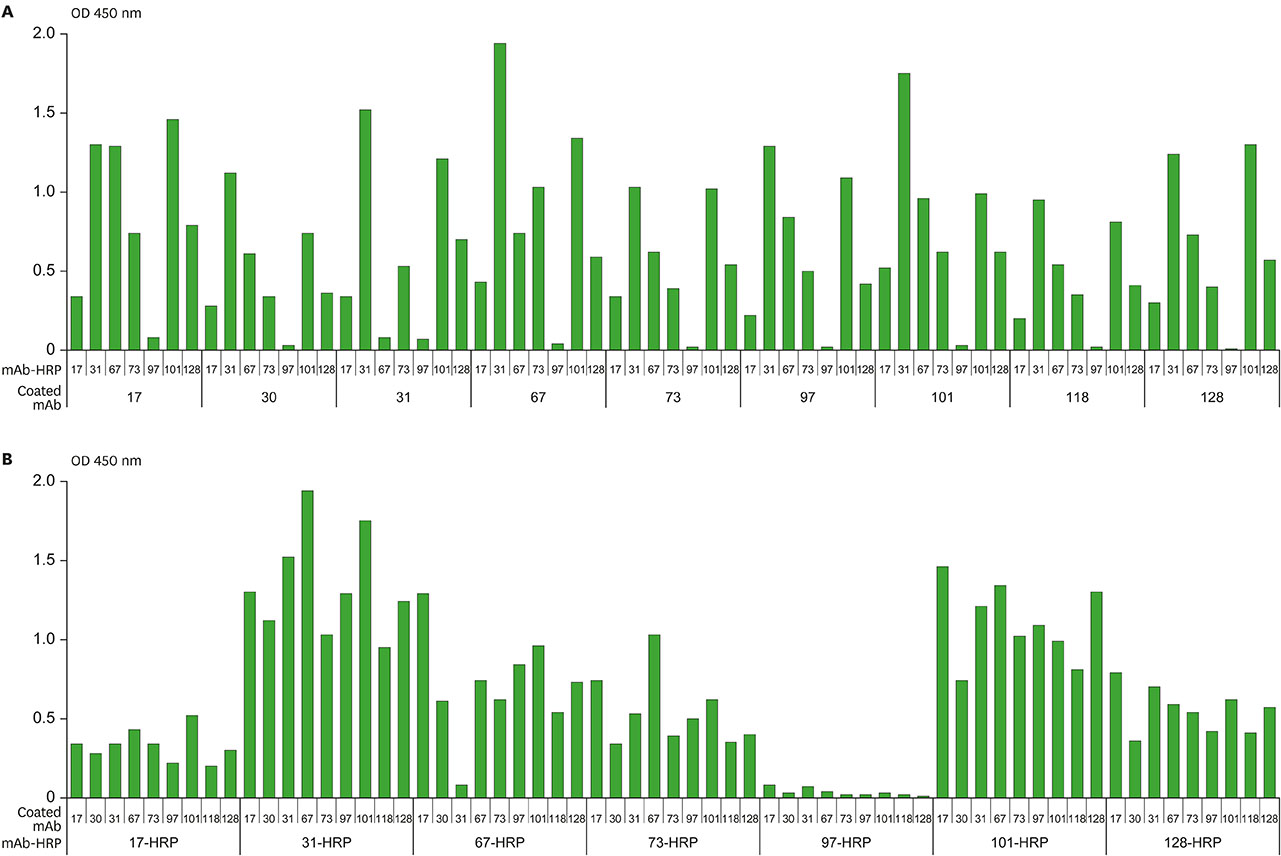
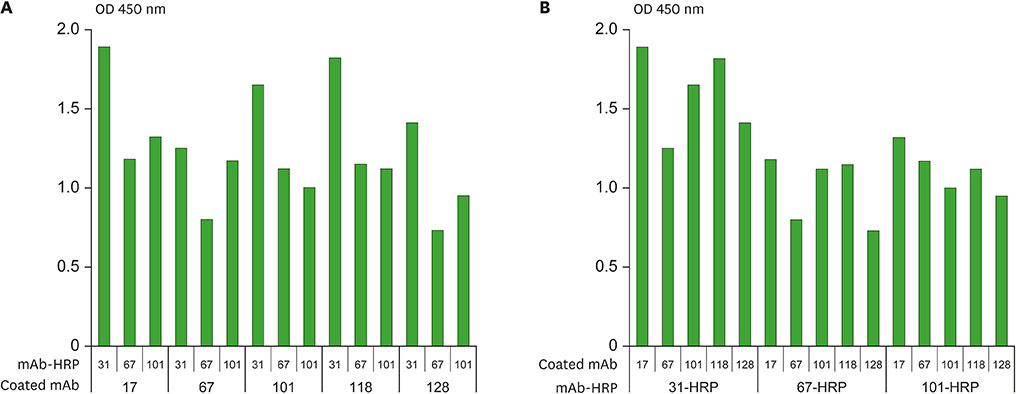
- Several studies have reported a good correlation between levels of serum hepatitis B virus surface antigen (HBsAg) and covalently closed circular DNA (cccDNA) before and after antiviral therapy. As a result, the quantification of HBsAg levels has attracted much attention in recent years as an important approach to evaluate viral activity. In this study, mAbs against HBsAg were generated and 9 mAbs (H17, H30, H31, H67, H73, H97, H101, H118, and H128) were investigated for optimization of HBsAg quantitation ELISA. Determination of the best combinations of mAbs for sandwich ELISA identified H17 and H31 mAbs as the ideal capture and horseradish peroxidase (HRP) conjugate mAbs, respectively. A standard curve for the current assay system exhibited linearity up to 40 ng/ml of HBsAg while a detection limit of approximately 1 ng/ml of HBsAg was also estimated, which was comparable to that of the other commercial ELISA kits. The ELISA system established in this study is particularly differentiated from other commercial kits in using mAbs for both capture and HRP conjugate, which provides a solution to inconsistency of quality and ethical issues in polyclonal antibodies production using laboratory animals.
MeSH Terms
Figure
Reference
-
1. Sonneveld MJ, Zoutendijk R, Janssen HL. Hepatitis B surface antigen monitoring and management of chronic hepatitis B. J Viral Hepat. 2011; 18:449–457.
Article2. Park H, Lee JM, Seo JH, Kim HS, Ahn SH, Kim DY, Han KH, Chon CY, Park JY. Predictive value of HBsAg quantification for determining the clinical course of genotype C HBeAg-negative carriers. Liver Int. 2012; 32:796–802.
Article3. Werle-Lapostolle B, Bowden S, Locarnini S, Wursthorn K, Petersen J, Lau G, Trepo C, Marcellin P, Goodman Z, Delaney WE 4th. Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology. 2004; 126:1750–1758.
Article4. Zoulim F. New insight on hepatitis B virus persistence from the study of intrahepatic viral cccDNA. J Hepatol. 2005; 42:302–308.
Article5. Chan HL, Wong VW, Tse AM, Tse CH, Chim AM, Chan HY, Wong GL, Sung JJ. Serum hepatitis B surface antigen quantitation can reflect hepatitis B virus in the liver and predict treatment response. Clin Gastroenterol Hepatol. 2007; 5:1462–1468.
Article6. Chuaypen N, Sriprapun M, Praianantathavorn K, Payungporn S, Wisedopas N, Poovorawan Y, Tangkijvanich P. Kinetics of serum HBsAg and intrahepatic cccDNA during pegylated interferon therapy in patients with HBeAg-positive and HBeAg-negative chronic hepatitis B. J Med Virol. 2017; 89:130–138.
Article7. Li J, Sun X, Fang J, Wang C, Han G, Ren W. Analysis of intrahepatic total HBV DNA, cccDNA and serum HBsAg level in chronic hepatitis B patients with undetectable serum HBV DNA during oral antiviral therapy. Clin Res Hepatol Gastroenterol. 2017.
Article8. Martinot-Peignoux M, Lapalus M, Asselah T, Marcellin P. HBsAg quantification: useful for monitoring natural history and treatment outcome. Liver Int. 2014; 34:Suppl 1. 97–107.
Article9. Karagoz E, Tanoglu A. Clinical usefulness of HBsAg quantification in patients with chronic hepatitis B infection. Hepat Mon. 2017; 17:e12293.
Article10. Hong KW, Kim CG, Lee SH, Chang KH, Shin YW, Ryoo KH, Kim SH, Kim YS. A novel anti-EGFR monoclonal antibody inhibiting tumor cell growth by recognizing different epitopes from cetuximab. J Biotechnol. 2010; 145:84–91.
Article11. Nakane PK, Kawaoi A. Peroxidase-labeled antibody. A new method of conjugation. J Histochem Cytochem. 1974; 22:1084–1091.
Article12. European Medicines Agency. Guideline on Bioanalytical Method Validation. London: European Medicines Agency;2011.13. Valentin MA, Ma S, Zhao A, Legay F, Avrameas A. Validation of immunoassay for protein biomarkers: bioanalytical study plan implementation to support pre-clinical and clinical studies. J Pharm Biomed Anal. 2011; 55:869–877.
Article14. Maylin S, Boyd A, Delaugerre C, Zoulim F, Lavocat F, Simon F, Girard PM, Lacombe K. Comparison between Elecsys HBsAg II and architect HBsAg QT assays for quantification of hepatitis B surface antigen among patients coinfected with HIV and hepatitis B virus. Clin Vaccine Immunol. 2012; 19:242–248.
Article15. Orito E, Fujiwara K, Kanie H, Ban T, Yamada T, Hayashi K. Quantitation of HBsAg predicts response to entecavir therapy in HBV genotype C patients. World J Gastroenterol. 2012; 18:5570–5575.
Article16. Chan HL, Thompson A, Martinot-Peignoux M, Piratvisuth T, Cornberg M, Brunetto MR, Tillmann HL, Kao JH, Jia JD, Wedemeyer H, et al. Hepatitis B surface antigen quantification: why and how to use it in 2011 - a core group report. J Hepatol. 2011; 55:1121–1131.
Article17. İnan N, Demirel A, Ünsur EK, Görmüs U, Sönmez E, Tabak F, Arisoy A. Comparison of chemiluminescence microparticle immunoassay and electrochemiluminescence immunoassay for detection of HBsAg. Viral Hepat J. 2014; 20:101–105.
Article18. Hadziyannis E. Quantification of HBsAg in serum: characteristics of the assays. OA Hepatol. 2013; 1:1–6.19. Andreotti PE, Ludwig GV, Peruski AH, Tuite JJ, Morse SS, Peruski LF Jr. Immunoassay of infectious agents. Biotechniques. 2003; 35:850–859.
Article20. Pohanka M. Monoclonal and polyclonal antibodies production – preparation of potent biorecognition element. J Appl Biomed. 2009; 7:115–121.
Article21. Cherlet M, Marc A. Stimulation of monoclonal antibody production of hybridoma cells by butyrate: evaluation of a feeding strategy and characterization of cell behaviour. Cytotechnology. 2000; 32:17–29.22. Gong X, Li D, Li X, Fang Q, Han X, Wu Y, Yang S, Shen BQ. Fed-batch culture optimization of a growth-associated hybridoma cell line in chemically defined protein-free media. Cytotechnology. 2006; 52:25–38.
Article23. Peng J, Song S, Xu L, Ma W, Liu L, Kuang H, Xu C. Development of a monoclonal antibody-based sandwich ELISA for peanut allergen Ara h 1 in food. Int J Environ Res Public Health. 2013; 10:2897–2905.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Eligibility Study of Anesthesia and Surgery for HBs Antigen Positive Patients
- Growing attention to an old marker, hepatitis B surface antigen, in the natural history of chronic hepatitis B
- Spontaneous Clearance of Hepatitis B Surface Antigenemia in a Hemodialysis Patient
- Prevalence of hepatitis B surface antigen in pediatric in patients
- Quantitative hepatitis B surface antigen predicts the antiviral response and hepatocellular carcinoma development in patients with chronic hepatitis B