Ann Lab Med.
2017 Jan;37(1):39-44. 10.3343/alm.2017.37.1.39.
Evaluation of BD MAX Staph SR Assay for Differentiating Between Staphylococcus aureus and Coagulase-Negative Staphylococci and Determining Methicillin Resistance Directly From Positive Blood Cultures
- Affiliations
-
- 1Department of Laboratory Medicine, College of Medicine, Seoul St. Mary's Hospital, Catholic University of Korea, Seoul, Korea. yjpk@catholic.ac.kr
- 2Department of Laboratory Medicine, College of Medicine, Uijeongbu St. Mary's Hospital, Catholic University of Korea, Uijeongbu, Korea.
- KMID: 2373612
- DOI: http://doi.org/10.3343/alm.2017.37.1.39
Abstract
- BACKGROUND
We evaluated the performance of the BD MAX StaphSR Assay (SR assay; BD, USA) for direct detection of Staphylococcus aureus and methicillin resistance not only in S. aureus but also in coagulase-negative Staphylococci (CNS) from positive blood cultures.
METHODS
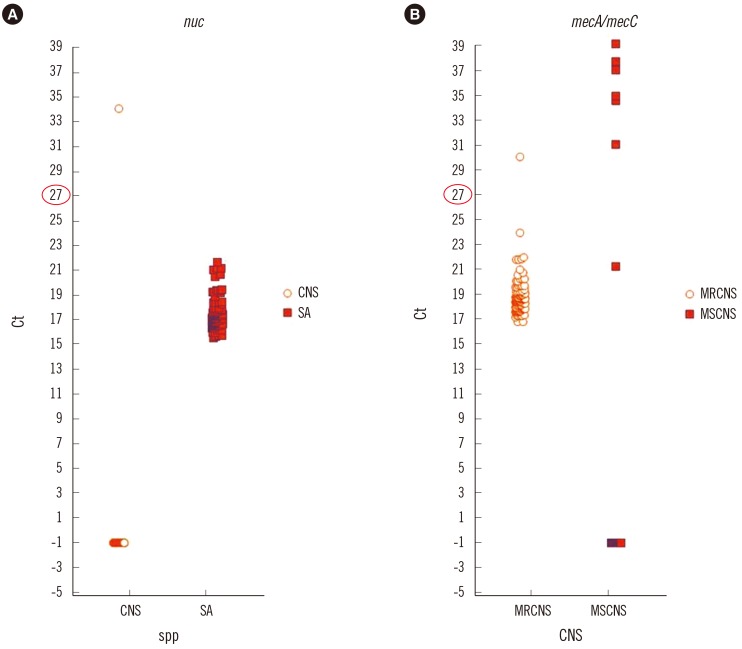
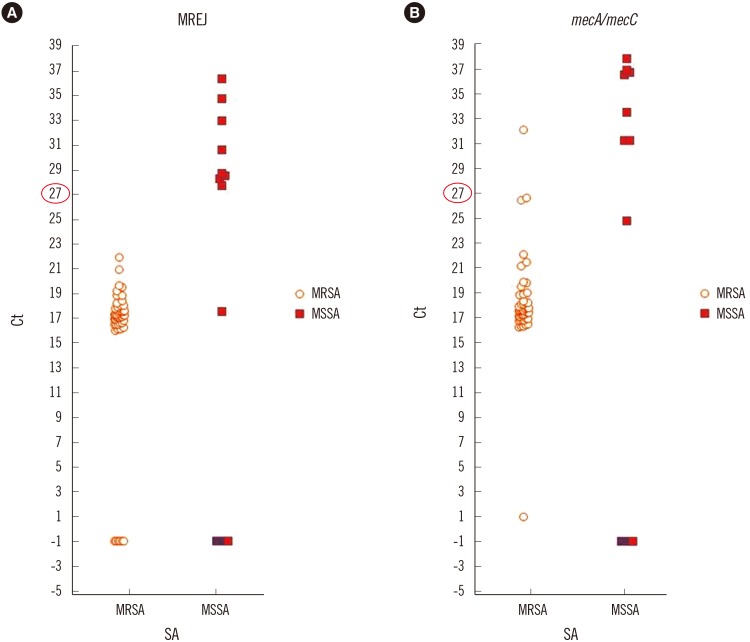
From 228 blood culture bottles, 103 S. aureus [45 methicillin-resistant S. aureus (MRSA), 55 methicillin-susceptible S. aureus (MSSA), 3 mixed infections (1 MRSA+Enterococcus faecalis, 1 MSSA+MRCNS, 1 MSSA+MSCNS)], and 125 CNS (102 MRCNS, 23 MSCNS) were identified by Vitek 2. For further analysis, we obtained the cycle threshold (Ct) values from the BD MAX system software to determine an appropriate cutoff value. For discrepancy analysis, conventional mecA/mecC PCR and oxacillin minimum inhibitory concentrations (MICs) were determined.
RESULTS
Compared to Vitek 2, the SR assay identified all 103 S. aureus isolates correctly but failed to detect methicillin resistance in three MRSA isolates. All 55 MSSA isolates were correctly identified by the SR assay. In the concordant cases, the highest Ct values for nuc, mecA, and mec right-extremity junction (MREJ) were 25.6, 22, and 22.2, respectively. Therefore, we selected Ct values from 0-27 as a range of positivity, and applying this cutoff, the sensitivity/specificity of the SR assay were 100%/100% for detecting S. aureus, and 97.9%/98.1% and 99.0%/95.8% for detecting methicillin resistance in S. aureus and CNS, respectively.
CONCLUSIONS
We propose a Ct cutoff value for nuc/mec assay without considering MREJ because mixed cultures of MSSA and MRCNS were very rare (0.4%) in the positive blood cultures.
Keyword
MeSH Terms
-
Anti-Bacterial Agents/pharmacology
Bacteremia/diagnosis/microbiology
Coagulase/metabolism
Humans
Methicillin-Resistant Staphylococcus aureus/drug effects/genetics/*isolation & purification
Microbial Sensitivity Tests
Oxacillin/pharmacology
Reagent Kits, Diagnostic
Staphylococcus/drug effects/enzymology/genetics/isolation & purification
Staphylococcus aureus/drug effects/genetics/*isolation & purification
Anti-Bacterial Agents
Coagulase
Oxacillin
Reagent Kits, Diagnostic
Figure
Reference
-
1. Snyder JW, Munier GK, Heckman SA, Camp P, Overman TL. Failure of the BD GeneOhm StaphSR assay for direct detection of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates in positive blood cultures collected in the United States. J Clin Microbiol. 2009; 47:3747–3748. PMID: 19794051.2. Savithri MB, Iyer V, Jones M, Yarwood T, Looke D, Kruger PS, et al. Epidemiology and significance of coagulase-negative staphylococci isolated in blood cultures from critically ill adult patients. Crit Care Resusc. 2011; 13:103–107. PMID: 21627578.3. Rogers KL, Fey PD, Rupp ME. Coagulase-negative staphylococcal infections. Infect Dis Clin North Am. 2009; 23:73–98. PMID: 19135917.4. Pasanen T, Korkeila M, Mero S, Tarkka E, Piiparinen H, Vnopio-Varkila J, et al. A selective broth enrichment combined with real-time nuc-mecA-PCR in the exclusion of MRSA. Apmis. 2010; 118:74–80. PMID: 20041874.BD MAX StaphSR [package insert]. Catalog no. 442999, publication P0153 (01). BD Diagnostics Sparks. MD, USA: 2013.6. CLSI. Performance standards for antimicrobial susceptibility testing. M100-S25. Wayne, PA: Clinical and Laboratory Standards Institute;2015.7. EURL-AR. European union reference laboratory antimicrobial resistance. Protocol for PCR amplification of MECA, MECC (MECALGA251), SPA and PVL. Lyngby, Denmark: DTU Food, National Food Institute;2012.8. Dalpke AH, Hofko M, Hamilton F, Meckenzie L, Zimmermann S, Templeton K. Evaluation of the BD Max StaphSR Assay for rapid identification of Staphylococcus aureus and Methicillin-resistant S. aureus in Positive Blood Culture Broths. J Clin Microbiol. 2015; 53:3630–3632. PMID: 26292311.9. Thomas L, van Hal S, O'Sullivan M, Kyme P, Iredell J. Failure of the BD GeneOhm StaphS/R assay for identification of Australian methicillin-resistant Staphylococcus aureus strains: duplex assays as the "gold standard" in settings of unknown SCCmec epidemiology. J Clin Microbiol. 2008; 46:4116–4117. PMID: 18945832.10. Chen FJ, Huang IW, Wang CH, Chen PC, Wang HY, Lai JF, et al. mecA-positive Staphylococcus aureus with low-level oxacillin MIC in Taiwan. J Clin Microbiol. 2012; 50:1679–1683. PMID: 22378906.11. Proulx MK, Palace SG, Gandra S, Torres B, Weir S, Stiles T, et al. Reversion From Methicillin Susceptibility to Methicillin Resistance in Staphylococcus aureus During Treatment of Bacteremia. J Infect Dis. 2016; 213:1041–1048. PMID: 26503983.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Rapid Detection of Staphylococcus aureus and Methicillin-Resistant S. aureus in Atopic Dermatitis by Using the BD Max StaphSR Assay
- Early Screening of Oxacillin-Resistant Staphylococcus aureus and Staphylococcus epidermidis from Blood Culture
- Differentiation of Staphylococcus aureus from Coagulase-Negative Staphylococci by Lipovitellin-Salt-Mannitol Agar
- Evaluation of Slide and Tube Coagulase Tests, and Latex Agglutination Kit for the Identification of Staphylcoccus aureus
- Evaluation of CHROMagar Staph aureus, a New Chromogenic Medium, for the Detection of Nosocomial Methicillin-Resistant Staphylococcus aureus