Ann Lab Med.
2016 Nov;36(6):536-541. 10.3343/alm.2016.36.6.536.
Prevalence of Major Methicillin-Resistant Staphylococcus aureus Clones in Korea Between 2001 and 2008
- Affiliations
-
- 1Division of Antimicrobial Resistance, Center for Infectious Disease, National Research Institute of Health, Centers for Disease Control and Prevention (KCDC), Cheongju, Korea. jeongok22@gmail.com
- KMID: 2373591
- DOI: http://doi.org/10.3343/alm.2016.36.6.536
Abstract
- BACKGROUND
Methicillin-resistant Staphylococcus aureus (MRSA) are important pathogens causing nosocomial infections in Korean hospitals. This study aimed to investigate the epidemiological and genetic diversity of clinical S. aureus isolates in healthcare settings from 2001 to 2008.
METHODS
Samples and data were obtained from 986 individuals as part of the National Antimicrobial Surveillance Project, involving 10 regions nationwide. Molecular typing studies, including multilocus sequence typing (MLST) and staphylococcal cassette chromosome mec (SCCmec) typing were performed, and a representative clone of Korean MRSA was classified by combinational grouping using a DiversiLab (DL; bioMérieux, France) repetitive element polymerase chain reaction (rep-PCR) system.
RESULTS
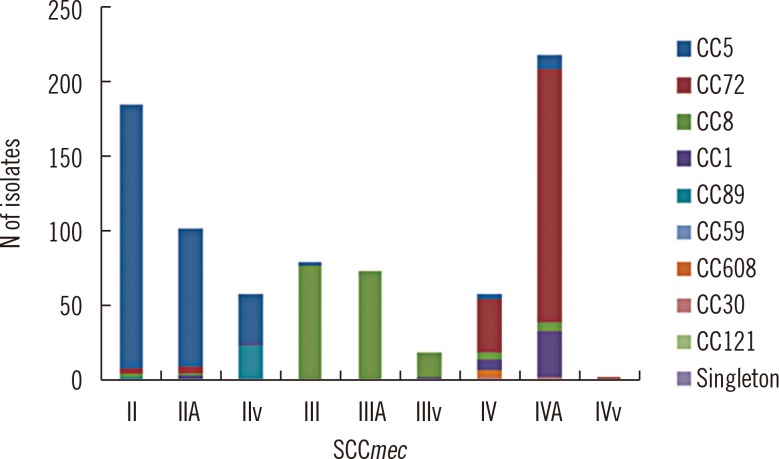
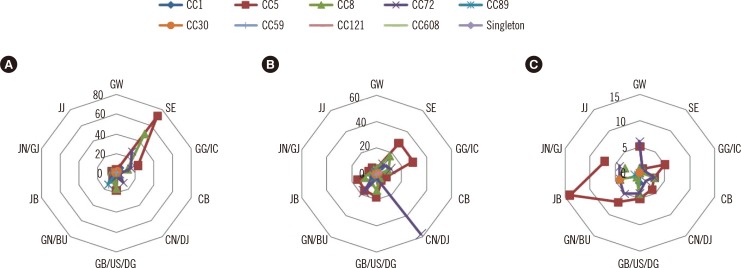
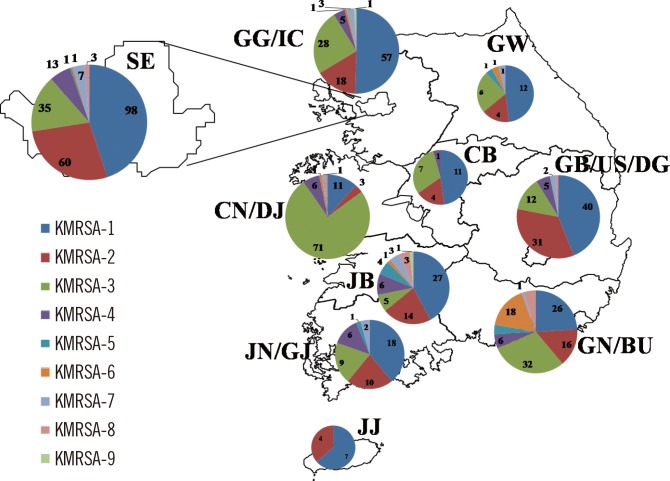
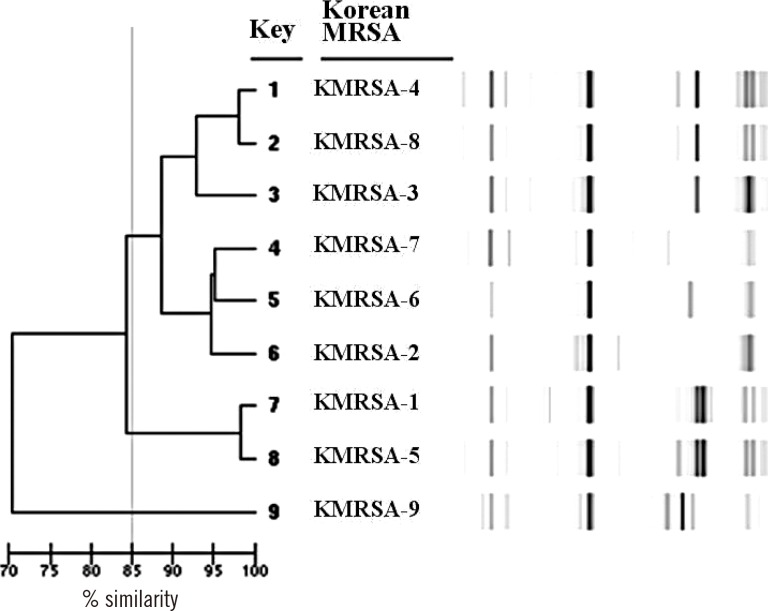
Nine Korean MRSA clones (KMRSA-1 to -9) were identified by analysis of genetic backgrounds and molecular characteristics. KMRSA-1 to -3, expressing clonal complex (CC) 5 (carrying SCCmec II), CC8 (carrying SCCmec III), and CC72 (carrying SCCmec IV) were spread nationwide. In contrast, KMRSA-6 was highly prevalent in Gyeongsangnam-do, and KMRSA-4 was highly prevalent in Jeollanam-do and Jeollabuk-do.
CONCLUSIONS
Epidemic KMRSA clones were genetically similar to major clones identified from the USA, with the exception of KMRSA-2, which had the SCCmec III type. Our results provide important insights into the distribution and molecular genetics of MRSA strains in Korea and may aid in the monitoring of MRSA spread throughout the country.
MeSH Terms
-
Bacterial Proteins/genetics
DNA, Bacterial/genetics/metabolism
Electrophoresis, Gel, Pulsed-Field
Hospitals
Humans
Methicillin-Resistant Staphylococcus aureus/*genetics/isolation & purification
Multilocus Sequence Typing
Multiplex Polymerase Chain Reaction
Prevalence
Republic of Korea/epidemiology
Staphylococcal Infections/diagnosis/*epidemiology/microbiology
Bacterial Proteins
DNA, Bacterial
Figure
Reference
-
1. Korea Centers for Disease Control and Prevention KCDC. Korean antimicrobial resistance monitoring system annual report. Cheongju, Korea: The Centers;2011.2. Larsen AR, Stegger M, Böcher S, Sørum M, Monnet DL, Skov RL. Emergence and characterization of community-associated methicillin-resistant Staphylococcus aureus infections in Denmark, 1999 to 2006. J Clin Microbiol. 2009; 47:73–78. PMID: 18971362.3. McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003; 41:5113–5120. PMID: 14605147.4. Johnson AP. Methicillin-resistant Staphylococcus aureus: the European landscape. J Antimicrob Chemother. 2011; 66(S4):iv43–iv48. PMID: 21521706.5. Kramer A, Wagenvoort H, Åhrén C, Daniels-Haardt I, Hartemann P, Kobayashi H, et al. Epidemiology of MRSA and current strategies in Europe and Japan. GMS Krankenhhyg Interdiszip. 2010; 5:Doc01. PMID: 20204100.6. Centers for Diseases Control and Prevention. Antibiotic resistance threats in the United States, 2013. Updated on Jul 2014. http://www.cdc.gov/drugresistance/threat-report-2013/.7. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Twenty-fourth informational supplement, M100-S24. Wayne, PA: Clinical and Laboratory Standards Institute;2014.8. Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002; 46:2155–2161. PMID: 12069968.9. Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999; 29:1128–1132. PMID: 10524952.10. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000; 38:1008–1015. PMID: 10698988.11. Shutt CK, Pounder JI, Page SR, Schaecher BJ, Woods GL. Clinical evaluation of the DiversiLab microbial typing system using repetitive-sequence-based PCR for characterization of Staphylococcus aureus strains. J Clin Microbiol. 2005; 43:1187–1192. PMID: 15750081.12. Tenover FC, Gay EA, Frye S, Eells SJ, Healy M, McGowan JE Jr. Comparison of typing results obtained for methicillin-resistant Staphylococcus aureus isolates with the DiversiLab system and pulsed-field gel electrophoresis. J Clin Microbiol. 2009; 47:2452–2457. PMID: 19553588.13. Köck R, Becker K, Cookson B, van Gemert-Pijnen JE, Harbarth S, Kluytmans J, et al. Methicillin-resistant Staphylococcus aureus (MRSA): burden of disease and control challenges in Europe. Euro Surveill. 2010; 15:19688. PMID: 20961515.14. Lee Y, Kim YA, Song W, Lee H, Lee HS, Jang SJ, et al. Recent trends in antimicrobial resistance in intensive care units in Korea. Korean J Nosocomial Infect Control. 2014; 19:29–36.15. Jones MB, Montgomery CP, Boyle-Vavra S, Shatzkes K, Maybank R, Frank BC, et al. Genomic and transcriptomic differences in community acquired methicillin resistant Staphylococcus aureus USA300 and USA400 strains. BMC Genomics. 2014; 15:1145. PMID: 25527145.16. Nimmo GR. USA300 abroad: global spread of a virulent strain of community associated methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2012; 18:725–734. PMID: 22448902.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Detection of Multidrug Resistant Patterns and Associated - genes of Methicillin Resistant Staphylococcus aureus ( MRSA ) Isolated from Clinical Specimens
- A statistical analysis of methicillin-resistant staphylococcus aureus
- A case of multiple furunculosis caused by methicillin-resistant staphylococcs aureus
- A Fatal Case of Infective Endocarditis Caused by Community-Associated Methicillin-Resistant Staphylococcus aureus ST72 in Korea
- Community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA)