Ann Lab Med.
2016 Nov;36(6):513-520. 10.3343/alm.2016.36.6.513.
Development and Characterization of Reference Materials for Genetic Testing: Focus on Public Partnerships
- Affiliations
-
- 1Centers for Disease Control and Prevention, Atlanta, GA, USA.
- 2Frederick National Laboratory for Cancer Research, National Cancer Institute, Gaithersburg, MD, USA.
- 3National Institute of Standards and Technology, Gaithersburg, MD, USA.
- 4Department of Laboratory Medicine, Dong-A University College of Medicine, Busan, Korea. jyhan@dau.ac.kr
- KMID: 2373588
- DOI: http://doi.org/10.3343/alm.2016.36.6.513
Abstract
- Characterized reference materials (RMs) are needed for clinical laboratory test development and validation, quality control procedures, and proficiency testing to assure their quality. In this article, we review the development and characterization of RMs for clinical molecular genetic tests. We describe various types of RMs and how to access and utilize them, especially focusing on the Genetic Testing Reference Materials Coordination Program (Get-RM) and the Genome in a Bottle (GIAB) Consortium. This review also reinforces the need for collaborative efforts in the clinical genetic testing community to develop additional RMs.
Keyword
MeSH Terms
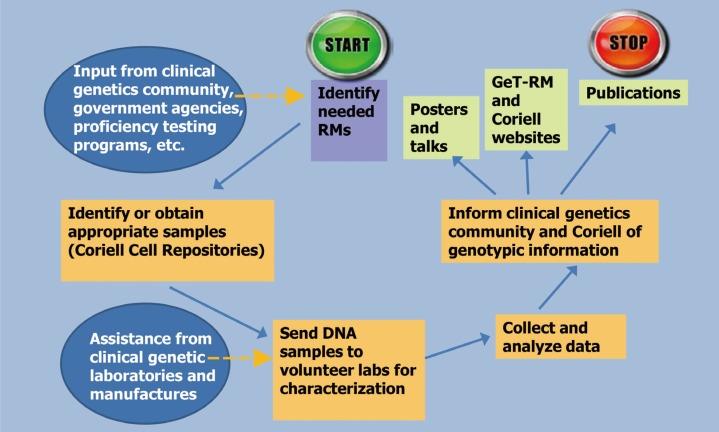
Figure
Reference
-
1. Kalman LV, Lubin IM, Barker S, du Sart D, Elles R, Grody WW, et al. Current landscape and new paradigms of proficiency testing and external quality assessment for molecular genetics. Arch Pathol Lab Med. 2013; 137:983–988. PMID: 23808472.2. Corbisier P, Pinheiro L, Mazoua S, Kortekaas AM, Chung PY, Gerganova T, et al. DNA copy number concentration measured by digital and droplet digital quantitative PCR using certified reference materials. Anal Bioanal Chem. 2015; 407:1831–1840. PMID: 25600685.3. Schrijver I, Aziz N, Farkas DH, Furtado M, Gonzalez AF, Greiner TC, et al. Opportunities and challenges associated with clinical diagnostic genome sequencing: a report of the Association for Molecular Pathology. J Mol Diagn. 2012; 14:525–540. PMID: 22918138.4. Gargis AS, Kalman L, Berry MW, Bick DP, Dimmock DP, Hambuch T, et al. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol. 2012; 30:1033–1036. PMID: 23138292.5. Chen B, Gagnon M, Shahangian S, Anderson NL, Howerton DA, Boone JD, et al. Good laboratory practices for molecular genetic testing for heritable diseases and conditions. MMWR Recomm Rep. 2009; 58(RR-6):1–37. PMID: 19521335.6. Chen B, O'Connell CD, Boone DJ, Amos JA, Beck JC, Chan MM, et al. Developing a sustainable process to provide quality control materials for genetic testing. Genet Med. 2005; 7:534–549. PMID: 16247292.7. Holden MJ, Madej RM, Minor P, Kalman LV. Molecular diagnostics: harmonization through reference materials, documentary standards and proficiency testing. Expert Rev Mol Diagn. 2011; 11:741–755. PMID: 21902536.8. National Institutes of Standards and Technology (NIST). RM 8398. Updated in 2015. https://www-s.nist.gov/srmors/view_detail.cfm?srm=8398.9. International Organization for Standardization. ISO Guide 30:2015- Reference materials- Selected terms and definitions. Updated on Feb 2015. https://www.iso.org/obp/ui/#iso:std:iso:guide:30:ed-3:v1:en.10. National Institute of Standards and Technology (NIST). http://www.nist.gov/srm/.11. European Reference Materials Consortium. European Reference Materials (ERM). Updated on Apr 2016. http://www.erm-crm.org/.12. World Health Organization. International reference materials. Updated in 2016. http://www.who.int/bloodproducts/ref_materials/en/.13. Genetic Testing Reference Material Coordination Program (Get-RM). Table of Higher Order Genetic Reference Materials. http://wwwn.cdc.gov/clia/Resources/GetRM/pdf/HigherOrderGeneticRMsTableWebsite.pdf.14. Joint Committee for Traceability in Laboratory Medicine (JCTLM). http://www.bipm.org/en/committees/jc/jctlm/.15. Zook JM, Chapman B, Wang J, Mittelman D, Hofmann O, Hide W, et al. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat Biotechnol. 2014; 32:246–251. PMID: 24531798.16. American College of Medical Genetics (ACMG). Standards and guidelines for clinical genetics laboratories, 2006 ed. Accessed on Apr 2015. http://www.acmg.net/Pages/ACMG_Activities/stds-2002/g.htm.17. Washington State Legislature. WAC 246-338-090 Quality control. Accessed on Apr 2015. http://app.leg.wa.gov/WAC/default.aspx?cite=246-338-090.18. College of American Pathologists (CAP). Accessed on Apr 2015. http://www.cap.org/apps/cap.portal.19. New York State. Clinical Laboratory Evaluation Program. Accessed on Apr 2015. http://www.wadsworth.org/clep.20. Clinical and Laboratory Standards Institute. Molecular methods for clinical genetics and oncology testing; approved guideline. 3rd ed. MM01-A3. Wayne, PA: Clinical and Laboratory Standards Institute;2012.21. Clinical and Laboratory Standards Institute. Verification and validation of multiplex nucleic acid assays; approved guideline. MM17-A. Wayne, PA: Clinical and Laboratory Standards Institute;2008.22. NCBI. Genetic Testing Registry. http://www.ncbi.nlm.nih.gov/gtr/.23. Centers for Disease Control and Prevention. Genetic Testing Reference Materials Coordination Program (GeT-RM). Updated on Mar 2015. http://wwwn.cdc.gov/clia/Resources/GetRM/.24. Coriell Institute. Coriell Cell Repositories. https://catalog.coriell.org/.25. Amos Wilson J, Pratt VM, Phansalkar A, Muralidharan K, Highsmith WE Jr, Beck JC, et al. Consensus characterization of 16 FMR1 reference materials: a consortium study. J Mol Diagn. 2008; 10:2–12. PMID: 18165276.26. Pratt VM, Caggana M, Bridges C, Buller AM, DiAntonio L, Highsmith WE, et al. Development of genomic reference materials for cystic fibrosis genetic testing. J Mol Diagn. 2009; 11:186–193. PMID: 19359498.27. Kalman L, Johnson MA, Beck J, Berry-Kravis E, Buller A, Casey B, et al. Development of genomic reference materials for Huntington disease genetic testing. Genet Med. 2007; 9:719–723. PMID: 18073586.28. Baker SD, Bale S, Booker J, Buller A, Das S, Friedman K, et al. Development and characterization of reference materials for MTHFR, SERPINA1, RET, BRCA1, and BRCA2 genetic testing. J Mol Diagn. 2009; 11:553–561. PMID: 19767587.29. Kalman L, Wilson JA, Buller A, Dixon J, Edelmann L, Geller L, et al. Development of genomic DNA reference materials for genetic testing of disorders common in people of Ashkenazi Jewish descent. J Mol Diagn. 2009; 11:530–536. PMID: 19815695.30. Kalman L, Leonard J, Gerry N, Tarleton J, Bridges C, Gastier-Foster JM, et al. Quality assurance for Duchenne and Becker muscular dystrophy genetic testing: development of a genomic DNA reference material panel. J Mol Diagn. 2011; 13:167–174. PMID: 21354051.31. Kalman L, Tarleton J, Hitch M, Hegde M, Hjelm N, Berry-Kravis E, et al. Development of a genomic DNA reference material panel for myotonic dystrophy type 1 (DM1) genetic testing. J Mol Diagn. 2013; 15:518–525. PMID: 23680132.32. Kalman LV, Tarleton JC, Percy AK, Aradhya S, Bale S, Barker SD, et al. Development of a genomic DNA reference material panel for Rett syndrome (MECP2-related disorders) genetic testing. J Mol Diagn. 2014; 16:273–279. PMID: 24508304.33. Pratt VM, Zehnbauer B, Wilson JA, Baak R, Babic N, Bettinotti M, et al. Characterization of 107 genomic DNA reference materials for CYP2D6, CYP2C19, CYP2C9, VKORC1, and UGT1A1: a GeT-RM and association for molecular pathology collaborative project. J Mol Diagn. 2010; 12:835–846. PMID: 20889555.34. Pratt VM, Everts RE, Aggarwal P, Beyer BN, Broeckel U, Epstein-Baak R, et al. Characterization of 137 genomic DNA reference materials for 28 pharmacogenetic genes: a Get-RM collaborative project. J Mol Diagn. 2016; 18:109–123. PMID: 26621101.35. Personal Genome Project: Harvard. http://www.personalgenomes.org/.36. Global Alliance for Genomics & Health. Benchmarking. Updated in 2016. https://genomicsandhealth.org/working-groups/benchmarking.37. Jenings L, Van Deerlin VM, Gulley ML. College of American Pathologists Molecular Pathology Resource Committee. Recommended principles and practices for validating clinical molecular pathology tests. Arch Pathol Lab Med. 2009; 133:743–755. PMID: 19415949.38. International Organization for Standardization. ISO 15189. Medical laboratories: requirements for quality and competence. Geneva: International Organization for Standardization;2012.39. Rehm HL, Bale SJ, Bayrak-Toydemir P, Berg JS, Brown KK, Deignan JL, et al. ACMG clinical laboratory standards for next-generation sequencing. Genet Med. 2013; 15:733–747. PMID: 23887774.40. NCBI. GeT-RM Browser. http://www.ncbi.nlm.nih.gov/variation/tools/get-rm.41. Illumina Platinum Genome. http://www.illumina.com/platinumgenomes/.42. Sims DJ, Harrington RD, Polley EC, Forbes TD, Mehaffey MG, McGregor PM 3rd, et al. Plasmid-based materials as multiplex quality controls and calibrators for clinical next-generation sequencing assays. J Mol Diagn. 2016; 18:336–349. PMID: 27105923.43. Wellcome Trust Sanger Institute. Catalogue of Somatic Mutations in Cancer (COSMIC). http://cancer.sanger.ac.uk/cosmic.44. Lih CJ, Sims DJ, Harrington RD, Polley EC, Zhao Y, Mehaffey MG, et al. Analytical validation and application of a targeted next-generation sequencing mutation-detection assay for use in treatment assignment in the NCI-MPACT trial. J Mol Diagn. 2016; 18:51–67. PMID: 26602013.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Direct-to-consumer genetic testing
- Genetic Testing and Genetic Counseling
- Current Status of Proficiency Testing Programs and Development of Reference Materials for Severe Acute Respiratory Syndrome Coronavirus 2 Molecular Diagnostic Tests
- Types, Production and Validation of Reference Materials for Viral Genetic Testing
- Regulated sandbox and disease-related genetic tests as a direct-to-consumer test