J Pathol Transl Med.
2017 Jan;51(1):24-31. 10.4132/jptm.2016.10.03.
KRAS Mutation Test in Korean Patients with Colorectal Carcinomas: A Methodological Comparison between Sanger Sequencing and a Real-Time PCR-Based Assay
- Affiliations
-
- 1Department of Hospital Pathology, College of Medicine, The Catholic University of Korea, Seoul, Korea. esjung@catholic.ac.kr
- KMID: 2367677
- DOI: http://doi.org/10.4132/jptm.2016.10.03
Abstract
- BACKGROUND
Mutations in the KRAS gene have been identified in approximately 50% of colorectal cancers (CRCs). KRAS mutations are well established biomarkers in anti-epidermal growth factor receptor therapy. Therefore, assessment of KRAS mutations is needed in CRC patients to ensure appropriate treatment.
METHODS
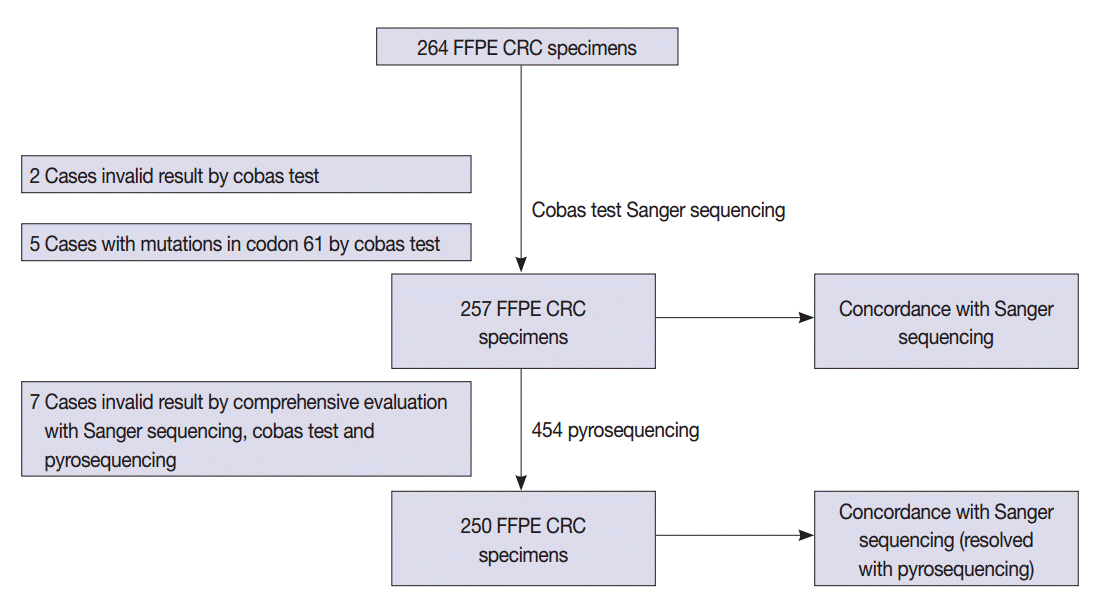
We compared the analytical performance of the cobas test to Sanger sequencing in 264 CRC cases. In addition, discordant specimens were evaluated by 454 pyrosequencing.
RESULTS
KRAS mutations for codons 12/13 were detected in 43.2% of cases (114/264) by Sanger sequencing. Of 257 evaluable specimens for comparison, KRAS mutations were detected in 112 cases (43.6%) by Sanger sequencing and 118 cases (45.9%) by the cobas test. Concordance between the cobas test and Sanger sequencing for each lot was 93.8% positive percent agreement (PPA) and 91.0% negative percent agreement (NPA) for codons 12/13. Results from the cobas test and Sanger sequencing were discordant for 20 cases (7.8%). Twenty discrepant cases were subsequently subjected to 454 pyrosequencing. After comprehensive analysis of the results from combined Sanger sequencing-454 pyrosequencing and the cobas test, PPA was 97.5% and NPA was 100%.
CONCLUSIONS
The cobas test is an accurate and sensitive test for detecting KRAS-activating mutations and has analytical power equivalent to Sanger sequencing. Prescreening using the cobas test with subsequent application of Sanger sequencing is the best strategy for routine detection of KRAS mutations in CRC.
Figure
Reference
-
1. Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003; 3:459–65.
Article2. Bos JL. Ras oncogenes in human cancer: a review. Cancer Res. 1989; 49:4682–9.3. Lee S, Brophy VH, Cao J, et al. Analytical performance of a PCR assay for the detection of KRAS mutations (codons 12/13 and 61) in formalin-fixed paraffin-embedded tissue samples of colorectal carcinoma. Virchows Arch. 2012; 460:141–9.4. Venook AP. Epidermal growth factor receptor-targeted treatment for advanced colorectal carcinoma. Cancer. 2005; 103:2435–46.
Article5. Dahabreh IJ, Terasawa T, Castaldi PJ, Trikalinos TA. Systematic review: anti-epidermal growth factor receptor treatment effect modification by KRAS mutations in advanced colorectal cancer. Ann Intern Med. 2011; 154:37–49.6. Querings S, Altmüller J, Ansén S, et al. Benchmarking of mutation diagnostics in clinical lung cancer specimens. PLoS One. 2011; 6:e19601.
Article7. Kim SY, Choi EJ, Yun JA, et al. Syndecan-1 expression is associated with tumor size and EGFR expression in colorectal carcinoma: a clinicopathological study of 230 cases. Int J Med Sci. 2015; 12:92–9.
Article8. Harlé A, Busser B, Rouyer M, et al. Comparison of COBAS 4800 KRAS, TaqMan PCR and high resolution melting PCR assays for the detection of KRAS somatic mutations in formalin-fixed paraffin embedded colorectal carcinomas. Virchows Arch. 2013; 462:329–35.9. Sharma A, Zhang G, Aslam S, Yu K, Chee M, Palma JF. Novel approach for clinical validation of the cobas KRAS mutation test in advanced colorectal cancer. Mol Diagn Ther. 2016; 20:231–40.10. Gonzalez de Castro D, Angulo B, Gomez B, et al. A comparison of three methods for detecting KRAS mutations in formalin-fixed colorectal cancer specimens. Br J Cancer. 2012; 107:345–51.11. Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008; 26:1626–34.12. Van Cutsem E, Köhne CH, Hitre E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009; 360:1408–17.
Article13. Bokemeyer C, Bondarenko I, Makhson A, et al. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the firstline treatment of metastatic colorectal cancer. J Clin Oncol. 2009; 27:663–71.
Article14. Douillard JY, Siena S, Cassidy J, et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol. 2010; 28:4697–705.
Article15. Malapelle U, Carlomagno C, Salatiello M, et al. KRAS mutation detection by high-resolution melting analysis significantly predicts clinical benefit of cetuximab in metastatic colorectal cancer. Br J Cancer. 2012; 107:626–31.16. Malapelle U, Carlomagno C, de Luca C, Bellevicine C, Troncone G. KRAS testing in metastatic colorectal carcinoma: challenges, controversies, breakthroughs and beyond. J Clin Pathol. 2014; 67:1–9.17. Angulo B, Lopez-Rios F, Gonzalez D. A new generation of companion diagnostics: cobas BRAF, KRAS and EGFR mutation detection tests. Expert Rev Mol Diagn. 2014; 14:517–24.18. Do H, Dobrovic A. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin Chem. 2015; 61:64–71.
Article19. Sah S, Chen L, Houghton J, et al. Functional DNA quantification guides accurate next-generation sequencing mutation detection in formalin-fixed, paraffin-embedded tumor biopsies. Genome Med. 2013; 5:77.
Article20. Tsiatis AC, Norris-Kirby A, Rich RG, et al. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications. J Mol Diagn. 2010; 12:425–32.21. Costello M, Pugh TJ, Fennell TJ, et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 2013; 41:e67.
Article22. Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011; 108:9530–5.
Article23. Xuan J, Yu Y, Qing T, Guo L, Shi L. Next-generation sequencing in the clinic: promises and challenges. Cancer Lett. 2013; 340:284–95.
Article24. Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008; 18:1851–8.
Article25. Quach N, Goodman MF, Shibata D. In vitro mutation artifacts after formalin fixation and error prone translesion synthesis during PCR. BMC Clin Pathol. 2004; 4:1.
Article26. Wong SQ, Li J, Tan AY, et al. Sequence artefacts in a prospective series of formalin-fixed tumours tested for mutations in hotspot regions by massively parallel sequencing. BMC Med Genomics. 2014; 7:23.
Article27. De Roock W, Jonker DJ, Di Nicolantonio F, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010; 304:1812–20.28. Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, et al. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007; 67:2643–8.
Article29. Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012; 486:532–6.30. Morris VK, Lucas FA, Overman MJ, et al. Clinicopathologic characteristics and gene expression analyses of non-KRAS 12/13, RAS-mutated metastatic colorectal cancer. Ann Oncol. 2014; 25:2008–14.31. Loupakis F, Ruzzo A, Cremolini C, et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer. 2009; 101:715–21.32. De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010; 11:753–62.33. Gao J, Li YY, Sun PN, Shen L. Comparative analysis of dideoxy sequencing, the KRAS StripAssay and pyrosequencing for detection of KRAS mutation. World J Gastroenterol. 2010; 16:4858–64.34. Angulo B, García-García E, Martínez R, et al. A commercial realtime PCR kit provides greater sensitivity than direct sequencing to detect KRAS mutations: a morphology-based approach in colorectal carcinoma. J Mol Diagn. 2010; 12:292–9.35. Whitehall V, Tran K, Umapathy A, et al. A multicenter blinded study to evaluate KRAS mutation testing methodologies in the clinical setting. J Mol Diagn. 2009; 11:543–52.36. Beau-Faller M, Degeorges A, Rolland E, et al. Cross-validation study for epidermal growth factor receptor and KRAS mutation detection in 74 blinded non-small cell lung carcinoma samples: a total of 5550 exons sequenced by 15 molecular French laboratories (evaluation of the EGFR mutation status for the administration of EGFR-TKIs in non-small cell lung carcinoma [ERMETIC] project- -part 1). J Thorac Oncol. 2011; 6:1006–15.37. Richman SD, Seymour MT, Chambers P, et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol. 2009; 27:5931–7.38. Gaedcke J, Grade M, Jung K, et al. KRAS and BRAF mutations in patients with rectal cancer treated with preoperative chemoradiotherapy. Radiother Oncol. 2010; 94:76–81.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Rapid and Sensitive Detection of KRAS Mutation by Peptide Nucleic Acid-based Real-time PCR Clamping: A Comparison with Direct Sequencing between Fresh Tissue and Formalin-fixed and Paraffin Embedded Tissue of Colorectal Cancer
- Comparison of Three BRAF Mutation Tests in Formalin-Fixed Paraffin Embedded Clinical Samples
- KRAS Mutation Detection in Non-small Cell Lung Cancer Using a Peptide Nucleic Acid-Mediated Polymerase Chain Reaction Clamping Method and Comparative Validation with Next-Generation Sequencing
- Mutational analysis of KRAS and its clinical implications in cervical cancer patients
- DNA Sequencing of p53 Gene Mutation in Colorectal Carcinomas