Chonnam Med J.
2017 Jan;53(1):56-63. 10.4068/cmj.2017.53.1.56.
Correlation between Infective Factors and Antibiotic Resistance in Enterococci Clinical Isolates in West of Iran
- Affiliations
-
- 1Department of Microbiology, Faculty of Medicine, Hamadan University of Medical Sciences, Hamadan, IR Iran. Mohammad.arabestani@gmail.com
- 2Brucellosis Research Center, Hamadan University of Medical Sciences, Hamadan, IR Iran.
- KMID: 2367340
- DOI: http://doi.org/10.4068/cmj.2017.53.1.56
Abstract
- The present study was done to scrutinize the possible relation between infective genes and antimicrobial resistance in Enterococcus faecalis and Enterococcus faecium. Considering the fact that the presence of recognized infective determinants among clinical isolates may promote the emergence of infections and persistence of Enterococci in hospital settings, which can lead to an increase in antimicrobial resistance. 175 E. faecalis and 67 E. faecium isolated from clinical specimens were used. The isolates were identified, and then antibiotic susceptibility testing was performed. The MIC of vancomycin and teicoplanin were determined by broth microdilution method. The presence of infective genes esp, hyl and asaâ‚ was scrutinized using PCR. Of the 280 enterococcal isolates, 175 (62.5%) isolates were identified as E. faecalis, 67 (24%) as E. faecium and 38 (13.5%) as Enterococcus spp. The results of the antibiotic susceptibility testing showed resistance rates of 5% and 73% to vancomycin and teicoplanin in E. faecalis and E. faecium isolates, respectively. The statistical analysis showed that the esp infective gene has significant associations with ciprofloxacin, erythromycin and tetracycline in E. faecium and with chloramphenicol in E. faecalis strains; the hyl with teicoplanin and vancomycin in E. faecium strains; and also asaâ‚ with vancomycin in E. faecium and with ampicillin and chloramphenicol in E. faecalis strains. Regarding the relationships between virulence genes and antibiotic resistance in strains of E. faecalis and E. faecium, detection of infective factors associated with invasive diseases has become a major issue of concern.
Keyword
MeSH Terms
-
Ampicillin
Anti-Bacterial Agents
Chloramphenicol
Ciprofloxacin
Drug Resistance, Microbial*
Enterococcus
Enterococcus faecalis
Enterococcus faecium
Erythromycin
Iran*
Methods
Polymerase Chain Reaction
Teicoplanin
Tetracycline
Vancomycin
Virulence
Ampicillin
Anti-Bacterial Agents
Chloramphenicol
Ciprofloxacin
Erythromycin
Teicoplanin
Tetracycline
Vancomycin
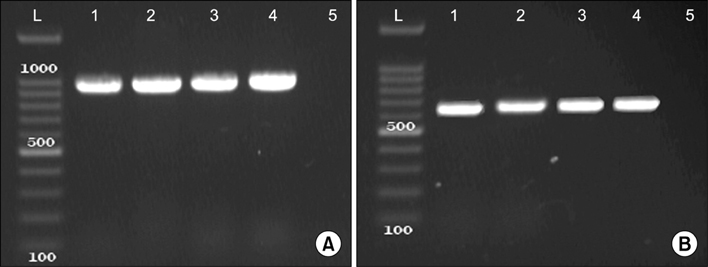
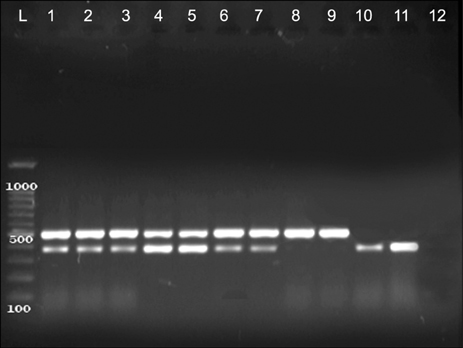
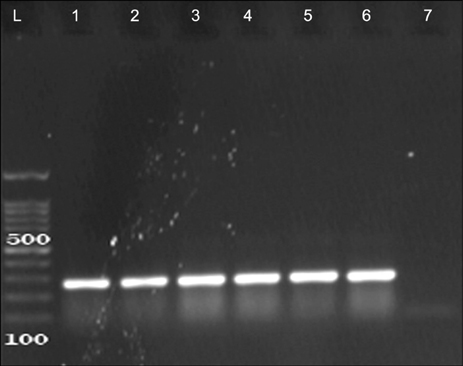
Figure
Reference
-
1. Nateghian AR, Ghasemi Ahari SM, Lahouti Harahdashti A, Navidnia M, Mehrazma M. Prevalence of vancomycin-resistant enterococci colonization, and susceptibility to linezolid in pediatric intensive care units of a referral pediatric center in Tehran, Iran. Arch Pediatr Infect Dis. 2014; 2:e16970.
Article2. Fernandes SC, Dhanashree B. Drug resistance & virulence determinants in clinical isolates of Enterococcus species. Indian J Med Res. 2013; 137:981–985.3. Mira MU, Deana M, Xora J, Vera G, Biljana M, Biljana R. Prevalence of different enterococcal species isolated from blood and their susceptibility to antimicrobial drugs in Vojvodina, Serbia, 2011-2013. Afr J Microbiol Res. 2014; 8:819–824.
Article4. Samadi Kafil H, Mohabati Mobarez A, Fourouzandeh Moghadam M. Multidrug resistant and most virulent Enterococcus faecium (strain 2653), isolated from hospitalized patient wound in Iran. Scholar J Med. 2012; 2:36–39.5. Van Tyne D, Gilmore MS. Friend turned foe: evolution of enterococcal virulence and antibiotic resistance. Annu Rev Microbiol. 2014; 68:337–356.
Article6. Feizabadi MM, Sayadi S, Shokrzadeh L, Parvin M, Yadegarynia D. Increase in prevalence of vancomycin resistant isolates of Enterococcous faecium at Labbafinejad hospital. Iran J Clin Infect Dis. 2008; 3:73–77.7. Medeiros AW, Pereira RI, Oliveira DV, Martins PD, d'Azevedo PA, Van der Sand S, et al. Molecular detection of virulence factors among food and clinical Enterococcus faecalis strains in South Brazil. Braz J Microbiol. 2014; 45:327–332.
Article8. Giridhara Upadhyaya PM, Umapathy BL, Ravikumar KL. Comparative study for the presence of enterococcal virulence factors gelatinase, hemolysin and biofilm among clinical and commensal isolates of enterococcus faecalis. J Lab Physicians. 2010; 2:100–104.
Article9. Biendo M, Adjidé C, Castelain S, Belmekki M, Rousseau F, Slama M, et al. Molecular characterization of glycopeptide-resistant enterococci from hospitals of the picardy region (france). Int J Microbiol. 2010; 2010:150464.
Article10. Vankerckhoven V, Huys G, Vancanneyt M, Snauwaert C, Swings J, Klare I, et al. Genotypic diversity, antimicrobial resistance, and virulence factors of human isolates and probiotic cultures constituting two intraspecific groups of Enterococcus faecium isolates. Appl Environ Microbiol. 2008; 74:4247–4255.
Article11. Camargo IL, Zanella RC, Gilmore MS, Darini AL. Virulence factors in vancomycin-resistant and vancomycin-susceptible Enterococcus faecalis from Brazil. Braz J Microbiol. 2008; 39:273–278.
Article12. Jahangiri S, Talebi M, Eslami G, Pourshafie MR. Prevalence of virulence factors and antibiotic resistance in vancomycin-resistant Enterococcus faecium isolated from sewage and clinical samples in Iran. Indian J Med Microbiol. 2010; 28:337–341.
Article13. Sharifi Y, Hasani A, Ghotaslou R, Varshochi M, Hasani A, Aghazadeh M, et al. Survey of Virulence Determinants among Vancomycin Resistant Enterococcus faecalis and Enterococcus faecium Isolated from Clinical Specimens of Hospitalized Patients of North west of Iran. Open Microbiol J. 2012; 6:34–39.
Article14. Manero A, Blanch AR. Identification of Enterococcus spp. with a biochemical key. Appl Environ Microbiol. 1999; 65:4425–4430.
Article15. Kariyama R, Mitsuhata R, Chow JW, Clewell DB, Kumon H. Simple and reliable multiplex PCR assay for surveillance isolates of vancomycin-resistant enterococci. J Clin Microbiol. 2000; 38:3092–3095.
Article16. Creti R, Imperi M, Bertuccini L, Fabretti F, Orefici G, Di Rosa R, et al. Survey for virulence determinants among Enterococcus faecalis isolated from different sources. J Med Microbiol. 2004; 53:13–20.
Article17. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility tests: approved standard - eleventh edition. CLSI document M02-A11. Wayne, PA: Clinical and Laboratory Standards Institute;2012.18. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: twenty-Third informational supplement. CLSI document M100-S23. Wayne, PA: Clinical and Laboratory Standards Institute;2013.19. Billström H, Lund B, Sullivan A, Nord CE. Virulence and antimicrobial resistance in clinical Enterococcus faecium. Int J Antimicrob Agents. 2008; 32:374–377.
Article20. Nowakiewicz A, Ziółkowska G, Zięba P, Trościańczyk A, Banach T, Kowalski C. Modified 16S-23S rRNA intergenic region restriction endonuclease analysis for species identification of Enterococcus strains isolated from pigs, compared with identification using classical methods and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Med Microbiol. 2015; 64:217–223.
Article21. Lavová M, Bezeková J, Čanigová M, Kročko M, Domig K. Species identification of Enterococci by biochemical tests and molecular-genetic methods. Potravinarstvo. 2014; 8:124–129.
Article22. Jurkovic D, Krizková L, Dusinský R, Belicová A, Sojka M, Krajcovic J, et al. Identification and characterization of enterococci from bryndza cheese. Lett Appl Microbiol. 2006; 42:553–559.
Article23. Sharifi Y, Hasani A, Ghotaslou R, Naghili B, Aghazadeh M, Milani M, et al. Virulence and antimicrobial resistance in enterococci isolated from urinary tract infections. Adv Pharm Bull. 2013; 3:197–201.24. Hällgren A, Claesson C, Saeedi B, Monstein HJ, Hanberger H, Nilsson LE. Molecular detection of aggregation substance, enterococcal surface protein, and cytolysin genes and in vitro adhesion to urinary catheters of Enterococcus faecalis and E. faecium of clinical origin. Int J Med Microbiol. 2009; 299:323–332.
Article25. Eaton TJ, Gasson MJ. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl Environ Microbiol. 2001; 67:1628–1635.
Article26. Eaton TJ, Gasson MJ. A variant enterococcal surface protein Esp(fm) in Enterococcus faecium; distribution among food, commensal, medical, and environmental isolates. FEMS Microbiol Lett. 2002; 216:269–275.
Article27. Vankerckhoven V, Van Autgaerden T, Vael C, Lammens C, Chapelle S, Rossi R, et al. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J Clin Microbiol. 2004; 42:4473–4479.
Article28. Shankar V, Baghdayan AS, Huycke MM, Lindahl G, Gilmore MS. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect Immun. 1999; 67:193–200.
Article29. Channaiah LH, Subramanyam B, McKinney LJ, Zurek L. Stored-product insects carry antibiotic-resistant and potentially virulent enterococci. FEMS Microbiol Ecol. 2010; 74:464–471.
Article30. Willems RJ, Homan W, Top J, van Santen-Verheuvel M, Tribe D, Manzioros X, et al. Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet. 2001; 357:853–855.
Article31. Woodford N, Soltani M, Hardy KJ. Frequency of esp in Enterococcus faecium isolates. Lancet. 2001; 358:584.
Article32. Sauer P, Síla J, Vágnerová I. Virulence factors in vancomycin-susceptible and vancomycin-resistant enterococci in the University Hospital Olomouc. Klin Mikrobiol Infekc Lek. 2009; 15:44–47.33. Camargo IL, Gilmore MS, Darini AL. Multilocus sequence typing and analysis of putative virulence factors in vancomycin-resistant and vancomycin-sensitive Enterococcus faecium isolates from Brazil. Clin Microbiol Infect. 2006; 12:1123–1130.
Article34. Worth LJ, Slavin MA, Vankerckhoven V, Goossens H, Grabsch EA, Thursky KA. Virulence determinants in vancomycin-resistant Enterococcus faecium vanB: clonal distribution, prevalence and significance of esp and hyl in Australian patients with haematological disorders. J Hosp Infect. 2008; 68:137–144.
Article35. Terkuran M, Erginkaya Z, Ünal E, Guran M, Kizilyildirim S, Gökce U, et al. The relationship between virulence factors and vancomycin resistance among Enterococci collected from food and human samples in Southern Turkey. Ankara Üniv Vet Fak Derg. 2014; 61:133–140.
Article36. Coque TM, Willems R, Cantón R, Del Campo R, Baquero F. High occurrence of esp among ampicillin-resistant and vancomycin-susceptible Enterococcus faecium clones from hospitalized patients. J Antimicrob Chemother. 2002; 50:1035–1038.
Article37. Leavis HL, Willems RJ, Top J, Spalburg E, Mascini EM, Fluit AC, et al. Epidemic and nonepidemic multidrug-resistant Enterococcus faecium. Emerg Infect Dis. 2003; 9:1108–1115.38. Comerlato CB, Resende MC, Caierão J, d'Azevedo PA. Presence of virulence factors in Enterococcus faecalis and Enterococcus faecium susceptible and resistant to vancomycin. Mem Inst Oswaldo Cruz. 2013; 108:590–595.
Article39. Kowalska-Krochmal B, Dworniczek E, Dolna I, Bania J, Wałecka E, Seniuk A, et al. Resistance patterns and occurrence of virulence determinants among GRE strains in southwestern Poland. Adv Med Sci. 2011; 56:304–310.
Article40. Rice LB, Carias L, Rudin S, Vael C, Goossens H, Konstabel C, et al. A potential virulence gene, hylEfm, predominates in Enterococcus faecium of clinical origin. J Infect Dis. 2003; 187:508–512.
Article41. Jankoska G, Trajkovska-Dokic E, Panovski N, Popovska-Jovanovska K, Petrovska M. Virulence factors and antibiotic resistance in Enterococcus faecalis isolated from urine samples. Prilozi. 2008; 29:57–66.42. Wardal E, Markowska K, Zabicka D, Wróblewska M, Giemza M, Mik E, et al. Molecular analysis of vanA outbreak of Enterococcus faecium in two Warsaw hospitals: the importance of mobile genetic elements. Biomed Res Int. 2014; 2014:575367.43. Baylan O, Nazik H, Bektöre B, Citil BE, Turan D, Ongen B, et al. The relationship between antibiotic resistance and virulence factors in urinary Enterococcus isolates. Mikrobiyol Bul. 2011; 45:430–445.44. Duprè I, Zanetti S, Schito AM, Fadda G, Sechi LA. Incidence of virulence determinants in clinical Enterococcus faecium and Enterococcus faecalis isolates collected in Sardinia (Italy). J Med Microbiol. 2003; 52:491–498.
Article45. Lund B, Edlund C. Bloodstream isolates of Enterococcus faecium enriched with the enterococcal surface protein gene, esp, show increased adhesion to eukaryotic cells. J Clin Microbiol. 2003; 41:5183–5185.
Article46. Van Wamel WJ, Hendrickx AP, Bonten MJ, Top J, Posthuma G, Willems RJ. Growth condition-dependent Esp expression by Enterococcus faecium affects initial adherence and biofilm formation. Infect Immun. 2007; 75:924–931.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Reduced Susceptibility to Biocides among Enterococci from Clinical and Non-Clinical Sources
- Survey for Correlation between Biofilm Formation and Virulence Determinants in a Collection of Pathogenic and Fecal Enterococcus faecalis Isolates
- Phenotypic Assays to Determine Virulence Factors of Uropathogenic
Escherichia coli (UPEC) Isolates and their Correlation with Antibiotic Resistance Pattern - Susceptibility of Fosfomycin against Vancomycin Resistant Enterococci
- Identification and Biochemical Reactions of Enterococci by a Simplified Identification System