Intest Res.
2015 Apr;13(2):112-121. 10.5217/ir.2015.13.2.112.
Epigenetic Alterations in Inflammatory Bowel Disease and Cancer
- Affiliations
-
- 1Research Institute, Dongnam Institute of Radiological & Medical Sciences (DIRAMS), Busan, Korea.
- 2Department of Internal Medicine, Haeundae Paik Hospital, Inje University College of Medicine, Busan, Korea. kto0440@paik.ac.kr
- KMID: 2284877
- DOI: http://doi.org/10.5217/ir.2015.13.2.112
Abstract
- Overwhelming evidences supports the idea that inflammatory bowel disease (IBD) is caused by a complex interplay between genetic alterations of multiple genes and an aberrant interaction with environmental factors. There is growing evidence that epigenetic factors can play a significant part in the pathogenesis of IBD. Significant effort has been invested in uncovering genetic and epigenetic factors, which may increase the risk of IBD, but progress has been slow, and few IBD-specific factors have been detected so far. It has been known for decades that DNA methylation is the most well studied epigenetic modification, and analysis of DNA methylation is leading to a new generation of cancer biomarkers. Therefore, in this review, we summarize the role of DNA methylation alteration in IBD pathogenesis, and discuss specific genes or genetic loci using recent molecular technology advances. Here, we suggest that DNA methylation should be studied in depth to understand the molecular pathways of IBD pathogenesis, and discuss epigenetic studies of IBD that may have a significant impact on the field of IBD research.
Figure
Cited by 2 articles
-
Lack of Aberrant Methylation in an Adjacent Area of Left-Sided Colorectal Cancer
Otgontuya Sambuudash, Hyun-Soo Kim, Mee Yon Cho
Yonsei Med J. 2017;58(4):749-755. doi: 10.3349/ymj.2017.58.4.749.Is there a potential role of fecal microbiota transplantation in the treatment of inflammatory bowel disease?
Chang Soo Eun
Intest Res. 2017;15(2):145-146. doi: 10.5217/ir.2017.15.2.145.
Reference
-
1. Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996; 87:159–170. PMID: 8861899.2. Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002; 16:6–21. PMID: 11782440.
Article3. Russo VEA, Martienssen RA, Riggs AD. Epigenetic mechanisms of gene regulation. New York: Cold Spring Harbor Laboratory Press;1996.4. Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999; 21:163–167. PMID: 9988266.5. Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003; 349:2042–2054. PMID: 14627790.
Article6. Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007; 8:286–298. PMID: 17339880.8. Chedin F, Lieber MR, Hsieh CL. The DNA methyltransferaselike protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc Natl Acad Sci U S A. 2002; 99:16916–16921. PMID: 12481029.9. Deplus R, Brenner C, Burgers WA, et al. Dnmt3L is a transcriptional repressor that recruits histone deacetylase. Nucleic Acids Res. 2002; 30:3831–3838. PMID: 12202768.
Article10. Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007; 128:683–692. PMID: 17320506.
Article11. Wajed SA, Laird PW, DeMeester TR. DNA methylation: an alternative pathway to cancer. Ann Surg. 2001; 234:10–20. PMID: 11420478.12. Gu M, Kim D, Bae Y, Choi J, Kim S, Song S. Analysis of microsatellite instability, protein expression and methylation status of hMLH1 and hMSH2 genes in gastric carcinomas. Hepatogastroenterology. 2009; 56:899–904. PMID: 19621725.13. Kane MF, Loda M, Gaida GM, et al. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997; 57:808–811. PMID: 9041175.14. Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996; 93:9821–9826. PMID: 8790415.
Article15. Esteller M, Silva JM, Dominguez G, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000; 92:564–569. PMID: 10749912.16. Merlo A, Herman JG, Mao L, et al. 5'CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995; 1:686–692. PMID: 7585152.17. Greger V, Passarge E, Hopping W, Messmer E, Horsthemke B. Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Hum Genet. 1989; 83:155–158. PMID: 2550354.
Article18. Chan TA, Glockner S, Yi JM, et al. Convergence of mutation and epigenetic alterations identifies common genes in cancer that predict for poor prognosis. PLoS Med. doi: 10.1371/journal.pmed.0050114. Published online 27 May 2008.
Article19. Schuebel KE, Chen W, Cope L, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007; 3:1709–1723. PMID: 17892325.
Article20. Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983; 301:89–92. PMID: 6185846.
Article21. Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002; 99:3740–3745. PMID: 11891299.23. Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003; 300:455. PMID: 12702868.
Article24. Rodriguez J, Frigola J, Vendrell E, et al. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006; 66:8462–9468. PMID: 16951157.
Article25. Fraga MF, Herranz M, Espada J, et al. A mouse skin multistage carcinogenesis model reflects the aberrant DNA methylation patterns of human tumors. Cancer Res. 2004; 64:5527–5534. PMID: 15313885.
Article26. Antequera F, Bird A. CpG islands. EXS. 1993; 64:169–185. PMID: 8418949.27. Lim DH, Maher ER. Genomic imprinting syndromes and cancer. Adv Genet. 2010; 70:145–175. PMID: 20920748.
Article28. Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010; 70:27–56. PMID: 20920744.
Article29. Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010; 28:573–621. PMID: 20192811.
Article30. Loftus EV Jr. Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences. Gastroenterology. 2004; 126:1504–1517. PMID: 15168363.
Article31. Thia KT, Loftus EV Jr. SandbornWJ. YangSK. An update on the epidemiology of inflammatory bowel disease in Asia. Am J Gastroenterol. 2008; 103:3167–3182. PMID: 19086963.32. Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001; 293:1068–1070. PMID: 11498573.
Article33. Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007; 448:427–434. PMID: 17653185.
Article34. Kim JM. Antimicrobial proteins in intestine and inflammatory bowel diseases. Intest Res. 2014; 12:20–33. PMID: 25349560.
Article35. Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012; 491:119–124. PMID: 23128233.36. Xavier RJ, Rioux JD. Genome-wide association studies: a new window into immune-mediated diseases. Nat Rev Immunol. 2008; 8:631–643. PMID: 18654571.37. Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008; 40:955–962. PMID: 18587394.38. Bird AP, Wolffe AP. Methylation-induced repression-belts, braces, and chromatin. Cell. 1999; 99:451–454. PMID: 10589672.
Article39. Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010; 465:721–727. PMID: 20535201.
Article40. Gloria L, Cravo M, Pinto A, et al. DNA hypomethylation and proliferative activity are increased in the rectal mucosa of patients with long-standing ulcerative colitis. Cancer. 1996; 78:2300–2306. PMID: 8940998.
Article41. Hsieh CJ, Klump B, Holzmann K, Borchard F, Gregor M, Porschen R. Hypermethylation of the p16INK4a promoter in colectomy specimens of patients with long-standing and extensive ulcerative colitis. Cancer Res. 1998; 58:3942–3945. PMID: 9731506.42. Tahara T, Shibata T, Nakamura M, et al. Effect of MDR1 gene promoter methylation in patients with ulcerative colitis. Int J Mol Med. 2009; 23:521–527. PMID: 19288029.
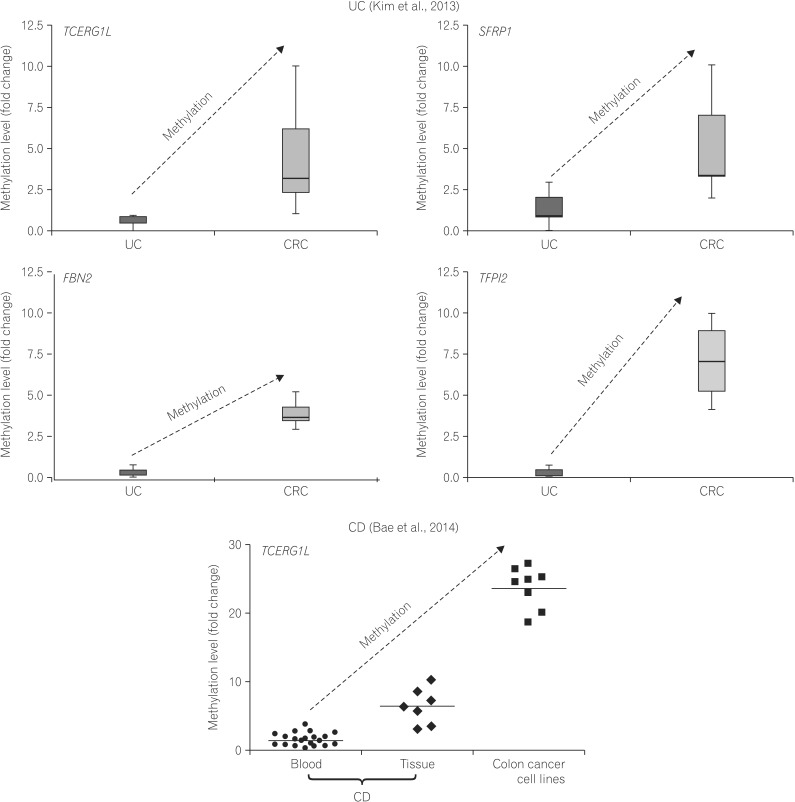
Article43. Kim TO, Park J, Kang MJ, et al. DNA hypermethylation of a selective gene panel as a risk marker for colon cancer in patients with ulcerative colitis. Int J Mol Med. 2013; 31:1255–1261. PMID: 23546389.
Article44. Lin Z, Hegarty JP, Cappel JA, et al. Identification of diseaseassociated DNA methylation in intestinal tissues from patients with inflammatory bowel disease. Clin Genet. 2011; 80:59–67. PMID: 20950376.
Article45. Cooke J, Zhang H, Greger L, et al. Mucosal genome-wide methylation changes in inflammatory bowel disease. Inflamm Bowel Dis. 2012; 18:2128–2137. PMID: 22419656.
Article46. Saito S, Kato J, Hiraoka S, et al. DNA methylation of colon mucosa in ulcerative colitis patients: correlation with inflammatory status. Inflamm Bowel Dis. 2011; 17:1955–1965. PMID: 21830274.47. Bae JH, Park J, Yang KM, Kim TO, Yi JM. Detection of DNA hypermethylation in sera of patients with Crohn's disease. Mol Med Rep. 2014; 9:725–729. PMID: 24317008.
Article48. Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000; 28:e32. PMID: 10734209.
Article49. Uhlmann K, Brinckmann A, Toliat MR, Ritter H, Nurnberg P. Evaluation of a potential epigenetic biomarker by quantitative methyl-single nucleotide polymorphism analysis. Electrophoresis. 2002; 23:4072–4079. PMID: 12481262.
Article50. Chin L, Gray JW. Translating insights from the cancer genome into clinical practice. Nature. 2008; 452:553–563. PMID: 18385729.
Article51. Caballero OL, Chen YT. Cancer/testis (CT) antigens: potential targets for immunotherapy. Cancer Sci. 2009; 100:2014–2021. PMID: 19719775.52. Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003; 3:253–266. PMID: 12671664.
Article53. Glockner SC, Dhir M, Yi JM, et al. Methylation of TFPI2 in stool DNA: a potential novel biomarker for the detection of colorectal cancer. Cancer Res. 2009; 69:4691–4699. PMID: 19435926.54. Lofton-Day C, Model F, Devos T, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem. 2008; 54:414–423. PMID: 18089654.
Article55. Brock MV, Hooker CM, Ota-Machida E, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008; 358:1118–1128. PMID: 18337602.
Article56. Yi JM, Dhir M, Van Neste L, et al. Genomic and epigenomic integration identifies a prognostic signature in colon cancer. Clin Cancer Res. 2011; 17:1535–1545. PMID: 21278247.57. Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999; 59:793–797. PMID: 10029064.58. Harris RA, Nagy-Szakal D, Pedersen N, et al. Genome-wide peripheral blood leukocyte DNA methylation microarrays identified a single association with inflammatory bowel diseases. Inflamm Bowel Dis. 2012; 18:2334–2341. PMID: 22467598.
Article59. Lin Z, Hegarty JP, Yu W, et al. Identification of disease-associated DNA methylation in B cells from Crohn's disease and ulcerative colitis patients. Dig Dis Sci. 2012; 57:3145–3153. PMID: 22821069.
Article60. Nimmo ER, Prendergast JG, Aldhous MC, et al. Genome-wide methylation profiling in Crohn's disease identifies altered epigenetic regulation of key host defense mechanisms including the Th17 pathway. Inflamm Bowel Dis. 2012; 18:889–899. PMID: 22021194.
Article61. Kim SW, Kim ES, Moon CM, Kim TI, Kim WH, Cheon JH. Abnormal genetic and epigenetic changes in signal transducer and activator of transcription 4 in the pathogenesis of inflammatory bowel diseases. Dig Dis Sci. 2012; 57:2600–2607. PMID: 22569826.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Pathogenesis and biomarkers of colorectal cancer by epigenetic alteration
- Recent Clinical Update of Acute Myeloid Leukemia: Focus on Epigenetic Therapies
- Genetic and Epigenetic Alterations in Bladder Cancer
- Effect of Helicobacter pylori Eradication on Epigenetic Changes in Gastric Cancer-related Genes
- Researches of Epigenetic Epidemiology for Infections and Radiation as Carcinogen