Allergy Asthma Immunol Res.
2010 Oct;2(4):215-227. 10.4168/aair.2010.2.4.215.
Unraveling the Genetic Basis of Asthma and Allergic Diseases
- Affiliations
-
- 1Department of Pediatric Immunology Research, Allergy Clinic Section, Children's Mercy Hospital & Clinics/School of Medicine, University of Missouri at Kansas City, Kansas City, USA. irosenwasser@cmh.edu
- KMID: 2260382
- DOI: http://doi.org/10.4168/aair.2010.2.4.215
Abstract
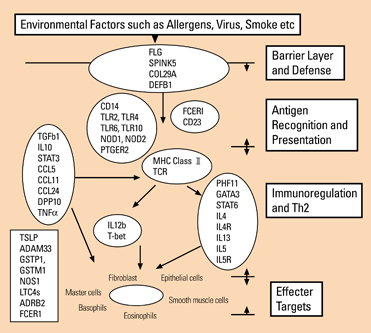
- Asthma and allergic diseases are believed to be complex genetic diseases which may result from the interaction of multiple genetic factors and environmental stimuli. In past decades, great efforts have been exerted in unraveling their genetic basis. The strategies in discovering genes and genetic variants, confirming their importance in pathogenesis of asthma and allergic diseases, as well as their strengths and limitations are summarized comprehensively and concisely. The current consensus about the genetic basis of asthma and allergic diseases is briefly described as well.
Keyword
Figure
Cited by 2 articles
-
Association between use of humidifier disinfectant and allergic rhinitis in Korean children: a cross-sectional study based on the eighth Panel Study on Korean Children (PSKC)
Tae Hwan Koh, Jong-Tae Park, Daeil Kwon, Kyeongmin Kwak
Ann Occup Environ Med. 2020;32:e9. doi: 10.35371/aoem.2020.32.e9.Association between use of humidifier disinfectant and allergic rhinitis in Korean children: a cross-sectional study based on the eighth Panel Study on Korean Children (PSKC)
Tae Hwan Koh, Jong-Tae Park, Daeil Kwon, Kyeongmin Kwak
Ann Occup Environ Med. 2020;32(1):. doi: 10.35371/aoem.2020.32.e9.
Reference
-
1. EPR3. Expert Panel Report 3 Guidelines for the Diagnosis and Management of Asthma. 2007. Bethesda, MD: US: Department of Health and Human services; National Institutes of Health; National Heart, Lung, nad Blood Institute; National Asthma Education and Prevention Program.2. Sandford A, Weir T, Pare P. The genetics of asthma. Am J Respir Crit Care Med. 1996. 153:1749–1765.3. Pinto LA, Stein RT, Kabesch M. Impact of genetics in childhood asthma. J Pediatr (Rio J). 2008. 84:S68–S75.4. Postma DS . Adkinson NF, Yunginger YJ, Busse WW, Bochner BS, Holgate ST, Simons FER, editors. Principles and application of genetics. Middleton's Allergy:Principles & Practice. 2003. 6th ed. Philadelphia, Pennsylvania 19106: Mosby;35–42.5. Khoury MJ, Beaty TH, Cohen BH. Genetic approaches to familial aggregation. III. linkage analysis. Fundamentals of Genetic Epidemiology. 1993. New York: Oxford University Press;284.6. Van Eerdewegh P, Little RD, Dupuis J, Del Mastro RG, Falls K, Simon J, Torrey D, Pandit S, McKenny J, Braunschweiger K, Walsh A, Liu Z, Hayward B, Folz C, Manning SP, Bawa A, Saracino L, Thackston M, Benchekroun Y, Capparell N, Wang M, Adair R, Feng Y, Dubois J, FitzGerald MG, Huang H, Gibson R, Allen KM, Pedan A, Danzig MR, Umland SP, Egan RW, Cuss FM, Rorke S, Clough JB, Holloway JW, Holgate ST, Keith TP. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2002. 418:426–430.7. Chavanas S, Bodemer C, Rochat A, Hamel-Teillac D, Ali M, Irvine AD, Bonafe JL, Wilkinson J, Taieb A, Barrandon Y, Harper JI, de Prost Y, Hovnanian A. Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nat Genet. 2000. 25:141–142.8. Balaci L, Spada MC, Olla N, Sole G, Loddo L, Anedda F, Naitza S, Zuncheddu MA, Maschio A, Altea D, Uda M, Pilia S, Sanna S, Masala M, Crisponi L, Fattori M, Devoto M, Doratiotto S, Rassu S, Mereu S, Giua E, Cadeddu NG, Atzeni R, Pelosi U, Corrias A, Perra R, Torrazza PL, Pirina P, Ginesu F, Marcias S, Schintu MG, Del Giacco GS, Manconi PE, Malerba G, Bisognin A, Trabetti E, Boner A, Pescollderungg L, Pignatti PF, Schlessinger D, Cao A, Pilia G. IRAK-M is involved in the pathogenesis of early-onset persistent asthma. Am J Hum Genet. 2007. 80:1103–1114.9. Allen M, Heinzmann A, Noguchi E, Abecasis G, Broxholme J, Ponting CP, Bhattacharyya S, Tinsley J, Zhang Y, Holt R, Jones EY, Lench N, Carey A, Jones H, Dickens NJ, Dimon C, Nicholls R, Baker C, Xue L, Townsend E, Kabesch M, Weiland SK, Carr D, von Mutius E, Adcock IM, Barnes PJ, Lathrop GM, Edwards M, Moffatt MF, Cookson WO. Positional cloning of a novel gene influencing asthma from chromosome 2q14. Nat Genet. 2003. 35:258–263.10. Laitinen T, Polvi A, Rydman P, Vendelin J, Pulkkinen V, Salmikangas P, Makela S, Rehn M, Pirskanen A, Rautanen A, Zucchelli M, Gullsten H, Leino M, Alenius H, Petays T, Haahtela T, Laitinen A, Laprise C, Hudson TJ, Laitinen LA, Kere J. Characterization of a common susceptibility locus for asthma-related traits. Science. 2004. 304:300–304.11. Nicolae D, Cox NJ, Lester LA, Schneider D, Tan Z, Billstrand C, Kuldanek S, Donfack J, Kogut P, Patel NM, Goodenbour J, Howard T, Wolf R, Koppelman GH, White SR, Parry R, Postma DS, Meyers D, Bleecker ER, Hunt JS, Solway J, Ober C. Fine mapping and positional candidate studies identify HLA-G as an asthma susceptibility gene on chromosome 6p21. Am J Hum Genet. 2005. 76:349–357.12. Zhang Y, Leaves NI, Anderson GG, Ponting CP, Broxholme J, Holt R, Edser P, Bhattacharyya S, Dunham A, Adcock IM, Pulleyn L, Barnes PJ, Harper JI, Abecasis G, Cardon L, White M, Burton J, Matthews L, Mott R, Ross M, Cox R, Moffatt MF, Cookson WO. Positional cloning of a quantitative trait locus on chromosome 13q14 that influences immunoglobulin E levels and asthma. Nat Genet. 2003. 34:181–186.13. Yoshinaka T, Nishii K, Yamada K, Sawada H, Nishiwaki E, Smith K, Yoshino K, Ishiguro H, Higashiyama S. Identification and characterization of novel mouse and human ADAM33s with potential metalloprotease activity. Gene. 2002. 282:227–236.14. Howard TD, Postma DS, Jongepier H, Moore WC, Koppelman GH, Zheng SL, Xu J, Bleecker ER, Meyers DA. Association of a disintegrin and metalloprotease 33 (ADAM33) gene with asthma in ethnically diverse populations. J Allergy Clin Immunol. 2003. 112:717–722.15. Walley AJ, Chavanas S, Moffatt MF, Esnouf RM, Ubhi B, Lawrence R, Wong K, Abecasis GR, Jones EY, Harper JI, Hovnanian A, Cookson WO. Gene polymorphism in Netherton and common atopic disease. Nat Genet. 2001. 29:175–178.16. Tan Z, Randall G, Fan J, Camoretti-Mercado B, Brockman-Schneider R, Pan L, Solway J, Gern JE, Lemanske RF, Nicolae D, Ober C. Allelespecific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet. 2007. 81:829–834.17. Marsh DG, Neely JD, Breazeale DR, Ghosh B, Freidhoff LR, Ehrlich-Kautzky E, Schou C, Krishnaswamy G, Beaty TH. Linkage analysis of IL4 and other chromosome 5q31.1 markers and total serum immunoglobulin E concentrations. Science. 1994. 264:1152–1156.18. Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996. 273:1516–1517.19. Ionita-Laza I, Rogers AJ, Lange C, Raby BA, Lee C. Genetic association analysis of copy-number variation (CNV) in human disease pathogenesis. Genomics. 2009. 93:22–26.20. Ober C, Hoffjan S. Asthma genetics 2006: the long and winding road to gene discovery. Genes Immun. 2006. 7:95–100.21. Hoffjan S, Nicolae D, Ober C. Association studies for asthma and atopic diseases: a comprehensive review of the literature. Respir Res. 2003. 4:14.22. Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008. 8:169–182.23. Steinke JW, Borish L, Rosenwasser LJ. 5. Genetics of hypersensitivity. J Allergy Clin Immunol. 2003. 111:S495–S501.24. Wu H, Romieu I, Shi M, Hancock DB, Li H, Sienra-Monge JJ, Chiu GY, Xu H, del Rio-Navarro BE, London SJ. Evaluation of candidate genes in a genome-wide association study of childhood asthma in Mexicans. J Allergy Clin Immunol. 2010. 125:321.e3–327.e3.25. Ragoussis J. Genotyping technologies for genetic research. Annu Rev Genomics Hum Genet. 2009. 10:117–133.26. Jorgenson E, Witte JS. Genome-wide association studies of cancer. Future Oncol. 2007. 3:419–427.27. Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E, Heinzmann A, Simma B, Frischer T, Willis-Owen SA, Wong KC, Illig T, Vogelberg C, Weiland SK, von Mutius E, Abecasis GR, Farrall M, Gut IG, Lathrop GM, Cookson WO. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007. 448:470–473.28. Sleiman PM, Annaiah K, Imielinski M, Bradfield JP, Kim CE, Frackelton EC, Glessner JT, Eckert AW, Otieno FG, Santa E, Thomas K, Smith RM, Glaberson W, Garris M, Gunnlaugsson S, Chiavacci RM, Allen J, Spergel J, Grundmeier R, Grunstein MM, Magnusson M, Bisgaard H, Grant SF, Hakonarson H. ORMDL3 variants associated with asthma susceptibility in North Americans of European ancestry. J Allergy Clin Immunol. 2008. 122:1225–1227.29. Galanter J, Choudhry S, Eng C, Nazario S, Rodriguez-Santana JR, Casal J, Torres-Palacios A, Salas J, Chapela R, Watson HG, Meade K, LeNoir M, Rodriguez-Cintron W, Avila PC, Burchard EG. ORMDL3 gene is associated with asthma in three ethnically diverse populations. Am J Respir Crit Care Med. 2008. 177:1194–1200.30. Leung TF, Sy HY, Ng MC, Chan IH, Wong GW, Tang NL, Waye MM, Lam CW. Asthma and atopy are associated with chromosome 17q21 markers in Chinese children. Allergy. 2009. 64:621–628.31. Verlaan DJ, Berlivet S, Hunninghake GM, Madore AM, Lariviere M, Moussette S, Grundberg E, Kwan T, Ouimet M, Ge B, Hoberman R, Swiatek M, Dias J, Lam KC, Koka V, Harmsen E, Soto-Quiros M, Avila L, Celedon JC, Weiss ST, Dewar K, Sinnett D, Laprise C, Raby BA, Pastinen T, Naumova AK. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet. 2009. 85:377–393.32. Ober C, Tan Z, Sun Y, Possick JD, Pan L, Nicolae R, Radford S, Parry RR, Heinzmann A, Deichmann KA, Lester LA, Gern JE, Lemanske RF Jr, Nicolae DL, Elias JA, Chupp GL. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med. 2008. 358:1682–1691.33. Himes BE, Hunninghake GM, Baurley JW, Rafaels NM, Sleiman P, Strachan DP, Wilk JB, Willis-Owen SA, Klanderman B, Lasky-Su J, Lazarus R, Murphy AJ, Soto-Quiros ME, Avila L, Beaty T, Mathias RA, Ruczinski I, Barnes KC, Celedon JC, Cookson WO, Gauderman WJ, Gilliland FD, Hakonarson H, Lange C, Moffatt MF, O'Connor GT, Raby BA, Silverman EK, Weiss ST. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet. 2009. 84:581–593.34. Spina D. PDE4 inhibitors: current status. Br J Pharmacol. 2008. 155:308–315.35. Hancock DB, Romieu I, Shi M, Sienra-Monge JJ, Wu H, Chiu GY, Li H, del Rio-Navarro BE, Willis-Owen SA, Weiss ST, Raby BA, Gao H, Eng C, Chapela R, Burchard EG, Tang H, Sullivan PF, London SJ. Genome-wide association study implicates chromosome 9q21.31 as a susceptibility locus for asthma in mexican children. PLoS Genet. 2009. 5:e1000623.36. Milili M, Gauthier L, Veran J, Mattei MG, Schiff C. A new Groucho TLE4 protein may regulate the repressive activity of Pax5 in human B lymphocytes. Immunology. 2002. 106:447–455.37. Yarmus M, Woolf E, Bernstein Y, Fainaru O, Negreanu V, Levanon D, Groner Y. Groucho/transducin-like Enhancer-of-split (TLE)-dependent and -independent transcriptional regulation by Runx3. Proc Natl Acad Sci U S A. 2006. 103:7384–7389.38. Weidinger S, Gieger C, Rodriguez E, Baurecht H, Mempel M, Klopp N, Gohlke H, Wagenpfeil S, Ollert M, Ring J, Behrendt H, Heinrich J, Novak N, Bieber T, Kramer U, Berdel D, von Berg A, Bauer CP, Herbarth O, Koletzko S, Prokisch H, Mehta D, Meitinger T, Depner M, von Mutius E, Liang L, Moffatt M, Cookson W, Kabesch M, Wichmann HE, Illig T. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet. 2008. 4:e1000166.39. Lee GR, Fields PE, Griffin TJ, Flavell RA. Regulation of the Th2 cytokine locus by a locus control region. Immunity. 2003. 19:145–153.40. Fields PE, Lee GR, Kim ST, Bartsevich VV, Flavell RA. Th2-specific chromatin remodeling and enhancer activity in the Th2 cytokine locus control region. Immunity. 2004. 21:865–876.41. Mathias RA, Grant AV, Rafaels N, Hand T, Gao L, Vergara C, Tsai YJ, Yang M, Campbell M, Foster C, Gao P, Togias A, Hansel NN, Diette G, Adkinson NF, Liu MC, Faruque M, Dunston GM, Watson HR, Bracken MB, Hoh J, Maul P, Maul T, Jedlicka AE, Murray T, Hetmanski JB, Ashworth R, Ongaco CM, Hetrick KN, Doheny KF, Pugh EW, Rotimi CN, Ford J, Eng C, Burchard EG, Sleiman PM, Hakonarson H, Forno E, Raby BA, Weiss ST, Scott AF, Kabesch M, Liang L, Abecasis G, Moffatt MF, Cookson WO, Ruczinski I, Beaty TH, Barnes KC. A genome-wide association study on African-ancestry populations for asthma. J Allergy Clin Immunol. 2010. 125:336.e4–346.e4.42. Sleiman PM, Flory J, Imielinski M, Bradfield JP, Annaiah K, Willis-Owen SA, Wang K, Rafaels NM, Michel S, Bonnelykke K, Zhang H, Kim CE, Frackelton EC, Glessner JT, Hou C, Otieno FG, Santa E, Thomas K, Smith RM, Glaberson WR, Garris M, Chiavacci RM, Beaty TH, Ruczinski I, Orange JM, Allen J, Spergel JM, Grundmeier R, Mathias RA, Christie JD, von Mutius E, Cookson WO, Kabesch M, Moffatt MF, Grunstein MM, Barnes KC, Devoto M, Magnusson M, Li H, Grant SF, Bisgaard H, Hakonarson H. Variants of DENND1B associated with asthma in children. N Engl J Med. 2010. 362:36–44.43. Del Villar K, Miller CA. Down-regulation of DENN/MADD, a TNF receptor binding protein, correlates with neuronal cell death in Alzheimer's disease brain and hippocampal neurons. Proc Natl Acad Sci U S A. 2004. 101:4210–4215.44. Lindstedt M, Lundberg K, Borrebaeck CA. Gene family clustering identifies functionally associated subsets of human in vivo blood and tonsillar dendritic cells. J Immunol. 2005. 175:4839–4846.45. Willinger T, Freeman T, Hasegawa H, McMichael AJ, Callan MF. Molecular signatures distinguish human central memory from effector memory CD8 T cell subsets. J Immunol. 2005. 175:5895–5903.46. Holloway JW, Koppelman GH. Identifying novel genes contributing to asthma pathogenesis. Curr Opin Allergy Clin Immunol. 2007. 7:69–74.47. Neale BM, Sham PC. The future of association studies: gene-based analysis and replication. Am J Hum Genet. 2004. 75:353–362.48. Wenzel SE. Asthma: defining of the persistent adult phenotypes. Lancet. 2006. 368:804–813.49. Kabesch M, Schedel M, Carr D, Woitsch B, Fritzsch C, Weiland SK, von Mutius E. IL-4/IL-13 pathway genetics strongly influence serum IgE levels and childhood asthma. J Allergy Clin Immunol. 2006. 117:269–274.50. Chan IH, Leung TF, Tang NL, Li CY, Sung YM, Wong GW, Wong CK, Lam CW. Gene-gene interactions for asthma and plasma total IgE concentration in Chinese children. J Allergy Clin Immunol. 2006. 117:127–133.51. Howard TD, Koppelman GH, Xu J, Zheng SL, Postma DS, Meyers DA, Bleecker ER. Gene-gene interaction in asthma: IL4RA and IL13 in a Dutch population with asthma. Am J Hum Genet. 2002. 70:230–236.52. Liu X, Beaty TH, Deindl P, Huang SK, Lau S, Sommerfeld C, Fallin MD, Kao WH, Wahn U, Nickel R. Associations between specific serum IgE response and 6 variants within the genes IL4, IL13, and IL4RA in German children: the German Multicenter Atopy Study. J Allergy Clin Immunol. 2004. 113:489–495.53. Ball TM, Castro-Rodriguez JA, Griffith KA, Holberg CJ, Martinez FD, Wright AL. Siblings, day-care attendance, and the risk of asthma and wheezing during childhood. N Engl J Med. 2000. 343:538–543.54. Illi S, von Mutius E, Lau S, Bergmann R, Niggemann B, Sommerfeld C, Wahn U. Early childhood infectious diseases and the development of asthma up to school age: a birth cohort study. BMJ. 2001. 322:390–395.55. Valdar W, Solberg LC, Gauguier D, Cookson WO, Rawlins JN, Mott R, Flint J. Genetic and environmental effects on complex traits in mice. Genetics. 2006. 174:959–984.56. Ober C, Thompson EE. Rethinking genetic models of asthma: the role of environmental modifiers. Curr Opin Immunol. 2005. 17:670–678.57. Martinez FD. CD14, endotoxin, and asthma risk: actions and interactions. Proc Am Thorac Soc. 2007. 4:221–225.58. Wang Z, Chen C, Niu T, Wu D, Yang J, Wang B, Fang Z, Yandava CN, Drazen JM, Weiss ST, Xu X. Association of asthma with beta(2)-adrenergic receptor gene polymorphism and cigarette smoking. Am J Respir Crit Care Med. 2001. 163:1404–1409.59. Eder W, Klimecki W, Yu L, von Mutius E, Riedler J, Braun-Fahrlander C, Nowak D, Martinez FD. Toll-like receptor 2 as a major gene for asthma in children of European farmers. J Allergy Clin Immunol. 2004. 113:482–488.60. Cameron L, Webster RB, Strempel JM, Kiesler P, Kabesch M, Ramachandran H, Yu L, Stern DA, Graves PE, Lohman IC, Wright AL, Halonen M, Klimecki WT, Vercelli D. Th2 cell-selective enhancement of human IL13 transcription by IL13-1112C>T, a polymorphism associated with allergic inflammation. J Immunol. 2006. 177:8633–8642.61. A genome-wide search for asthma susceptibility loci in ethnically diverse populations. The Collaborative Study on the Genetics of Asthma (CSGA). Nat Genet. 1997. 15:389–392.62. Xu J, Meyers DA, Ober C, Blumenthal MN, Mellen B, Barnes KC, King RA, Lester LA, Howard TD, Solway J, Langefeld CD, Beaty TH, Rich SS, Bleecker ER, Cox NJ. Genomewide screen and identification of gene-gene interactions for asthma-susceptibility loci in three U.S. populations: collaborative study on the genetics of asthma. Am J Hum Genet. 2001. 68:1437–1446.63. Scirica CV, Celedon JC. Genetics of asthma: potential implications for reducing asthma disparities. Chest. 2007. 132:770S–781S.64. Mak JC, Leung HC, Ho SP, Law BK, Ho AS, Lam WK, Ip MS, Chan-Yeung MM. Analysis of TGF-beta(1) gene polymorphisms in Hong Kong Chinese patients with asthma. J Allergy Clin Immunol. 2006. 117:92–96.65. Kim SH, Park HS, Holloway JW, Shin HD, Park CS. Association between a TGFbeta1 promoter polymorphism and rhinosinusitis in aspirin-intolerant asthmatic patients. Respir Med. 2007. 101:490–495.66. Stephens JC, Schneider JA, Tanguay DA, Choi J, Acharya T, Stanley SE, Jiang R, Messer CJ, Chew A, Han JH, Duan J, Carr JL, Lee MS, Koshy B, Kumar AM, Zhang G, Newell WR, Windemuth A, Xu C, Kalbfleisch TS, Shaner SL, Arnold K, Schulz V, Drysdale CM, Nandabalan K, Judson RS, Ruano G, Vovis GF. Haplotype variation and linkage disequilibrium in 313 human genes. Science. 2001. 293:489–493.67. Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. Genetic structure of human populations. Science. 2002. 298:2381–2385.68. Drake KA, Galanter JM, Burchard EG. Race, ethnicity and social class and the complex etiologies of asthma. Pharmacogenomics. 2008. 9:453–462.69. McHugh MK, Symanski E, Pompeii LA, Delclos GL. Prevalence of asthma among adult females and males in the United States: results from the National Health and Nutrition Examination Survey (NHANES), 2001-2004. J Asthma. 2009. 46:759–766.70. Meng JF, McFall C, Rosenwasser LJ. Polymorphism R62W results in resistance of CD23 to enzymatic cleavage in cultured cells. Genes Immun. 2007. 8:215–223.71. Hummelshoj T, Bodtger U, Datta P, Malling HJ, Oturai A, Poulsen LK, Ryder LP, Sorensen PS, Svejgaard E, Svejgaard A. Association between an interleukin-13 promoter polymorphism and atopy. Eur J Immunogenet. 2003. 30:355–359.72. Heinzmann A, Mao XQ, Akaiwa M, Kreomer RT, Gao PS, Ohshima K, Umeshita R, Abe Y, Braun S, Yamashita T, Roberts MH, Sugimoto R, Arima K, Arinobu Y, Yu B, Kruse S, Enomoto T, Dake Y, Kawai M, Shimazu S, Sasaki S, Adra CN, Kitaichi M, Inoue H, Yamauchi K, Tomichi N, Kurimoto F, Hamasaki N, Hopkin JM, Izuhara K, Shirakawa T, Deichmann KA. Genetic variants of IL-13 signalling and human asthma and atopy. Hum Mol Genet. 2000. 9:549–559.73. Arima K, Umeshita-Suyama R, Sakata Y, Akaiwa M, Mao XQ, Enomoto T, Dake Y, Shimazu S, Yamashita T, Sugawara N, Brodeur S, Geha R, Puri RK, Sayegh MH, Adra CN, Hamasaki N, Hopkin JM, Shirakawa T, Izuhara K. Upregulation of IL-13 concentration in vivo by the IL13 variant associated with bronchial asthma. J Allergy Clin Immunol. 2002. 109:980–987.74. Kiesler P, Shakya A, Tantin D, Vercelli D. An allergy-associated polymorphism in a novel regulatory element enhances IL13 expression. Hum Mol Genet. 2009. 18:4513–4520.75. Cookson W. The immunogenetics of asthma and eczema: a new focus on the epithelium. Nat Rev Immunol. 2004. 4:978–988.76. Hudson TJ. Skin barrier function and allergic risk. Nat Genet. 2006. 38:399–400.77. Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, Goudie DR, Sandilands A, Campbell LE, Smith FJ, O'Regan GM, Watson RM, Cecil JE, Bale SJ, Compton JG, DiGiovanna JJ, Fleckman P, Lewis-Jones S, Arseculeratne G, Sergeant A, Munro CS, El Houate B, McElreavey K, Halkjaer LB, Bisgaard H, Mukhopadhyay S, McLean WH. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006. 38:441–446.78. Soderhall C, Marenholz I, Kerscher T, Ruschendorf F, Esparza-Gordillo J, Worm M, Gruber C, Mayr G, Albrecht M, Rohde K, Schulz H, Wahn U, Hubner N, Lee YA. Variants in a novel epidermal collagen gene (COL29A1) are associated with atopic dermatitis. PLoS Biol. 2007. 5:e242.79. Levy H, Raby BA, Lake S, Tantisira KG, Kwiatkowski D, Lazarus R, Silverman EK, Richter B, Klimecki WT, Vercelli D, Martinez FD, Weiss ST. Association of defensin beta-1 gene polymorphisms with asthma. J Allergy Clin Immunol. 2005. 115:252–258.80. Kim E, Lee JE, Namkung JH, Kim PS, Kim S, Shin ES, Cho EY, Yang JM. Single nucleotide polymorphisms and the haplotype in the DEFB1 gene are associated with atopic dermatitis in a Korean population. J Dermatol Sci. 2009. 54:25–30.81. Kumar H, Kawai T, Akira S. Toll-like receptors and innate immunity. Biochem Biophys Res Commun. 2009. 388:621–625.82. Chen G, Shaw MH, Kim YG, Nunez G. NOD-like receptors: role in innate immunity and inflammatory disease. Annu Rev Pathol. 2009. 4:365–398.83. Hizawa N, Yamaguchi E, Jinushi E, Kawakami Y. A common FCER1B gene promoter polymorphism influences total serum IgE levels in a Japanese population. Am J Respir Crit Care Med. 2000. 161:906–909.84. Park HW, Shin ES, Lee JE, Kim SH, Kim SS, Chang YS, Kim YK, Min KU, Kim YY, Cho SH. Association between genetic variations in prostaglandin E2 receptor subtype EP3 gene (Ptger3) and asthma in the Korean population. Clin Exp Allergy. 2007. 37:1609–1615.85. Donfack J, Tsalenko A, Hoki DM, Parry R, Solway J, Lester LA, Ober C. HLA-DRB1*01 alleles are associated with sensitization to cockroach allergens. J Allergy Clin Immunol. 2000. 105:960–966.86. Moffatt MF, Hill MR, Cornelis F, Schou C, Faux JA, Young RP, James AL, Ryan G, le Souef P, Musk AW, Hopkin JM. Genetic linkage of T-cell receptor alpha/delta complex to specific IgE responses. Lancet. 1994. 343:1597–1600.87. Smit LA, Siroux V, Bouzigon E, Oryszczyn MP, Lathrop M, Demenais F, Kauffmann F. CD14 and toll-like receptor gene polymorphisms, country living, and asthma in adults. Am J Respir Crit Care Med. 2009. 179:363–368.88. Kormann MS, Depner M, Hartl D, Klopp N, Illig T, Adamski J, Vogelberg C, Weiland SK, von Mutius E, Kabesch M. Toll-like receptor heterodimer variants protect from childhood asthma. J Allergy Clin Immunol. 2008. 122:86–92. e1–e8.89. Weidinger S, Klopp N, Rummler L, Wagenpfeil S, Novak N, Baurecht HJ, Groer W, Darsow U, Heinrich J, Gauger A, Schafer T, Jakob T, Behrendt H, Wichmann HE, Ring J, Illig T. Association of NOD1 polymorphisms with atopic eczema and related phenotypes. J Allergy Clin Immunol. 2005. 116:177–184.90. Eder W, Klimecki W, Yu L, von Mutius E, Riedler J, Braun-Fahrlander C, Nowak D, Holst O, Martinez FD. Association between exposure to farming, allergies and genetic variation in CARD4/NOD1. Allergy. 2006. 61:1117–1124.91. Traidl-Hoffmann C, Mariani V, Hochrein H, Karg K, Wagner H, Ring J, Mueller MJ, Jakob T, Behrendt H. Pollen-associated phytoprostanes inhibit dendritic cell interleukin-12 production and augment T helper type 2 cell polarization. J Exp Med. 2005. 201:627–636.92. Tantisira KG, Silverman ES, Mariani TJ, Xu J, Richter BG, Klanderman BJ, Litonjua AA, Lazarus R, Rosenwasser LJ, Fuhlbrigge AL, Weiss ST. FCER2: a pharmacogenetic basis for severe exacerbations in children with asthma. J Allergy Clin Immunol. 2007. 120:1285–1291.93. Larche M, Robinson DS, Kay AB. The role of T lymphocytes in the pathogenesis of asthma. J Allergy Clin Immunol. 2003. 111:450–463. quiz 64.94. Pulendran B, Palucka K, Banchereau J. Sensing pathogens and tuning immune responses. Science. 2001. 293:253–256.95. Minelli C, Granell R, Newson R, Rose-Zerilli MJ, Torrent M, Ring SM, Holloway JW, Shaheen SO, Henderson JA. Glutathione-S-transferase genes and asthma phenotypes: a Human Genome Epidemiology (HuGE) systematic review and meta-analysis including unpublished data. Int J Epidemiol. 2010. 39:539–562.96. Gao PS, Kawada H, Kasamatsu T, Mao XQ, Roberts MH, Miyamoto Y, Yoshimura M, Saitoh Y, Yasue H, Nakao K, Adra CN, Kun JF, Moro-oka S, Inoko H, Ho LP, Shirakawa T, Hopkin JM. Variants of NOS1, NOS2, and NOS3 genes in asthmatics. Biochem Biophys Res Commun. 2000. 267:761–763.97. Sayers I, Barton S, Rorke S, Beghe B, Hayward B, Van Eerdewegh P, Keith T, Clough JB, Ye S, Holloway JW, Sampson AP, Holgate ST. Allelic association and functional studies of promoter polymorphism in the leukotriene C4 synthase gene (LTC4S) in asthma. Thorax. 2003. 58:417–424.98. Sharma S, Sharma A, Kumar S, Sharma SK, Ghosh B. Association of TNF haplotypes with asthma, serum IgE levels, and correlation with serum TNF-alpha levels. Am J Respir Cell Mol Biol. 2006. 35:488–495.99. Lee JH, Moore JH, Park SW, Jang AS, Uh ST, Kim YH, Park CS, Park BL, Shin HD. Genetic interactions model among Eotaxin gene polymorphisms in asthma. J Hum Genet. 2008. 53:867–875.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Overview and challenges of current genetic research on allergic diseases in Korean children
- Genetic studies of allergic disease and their clinical implications
- Genetic predisposition for atopy and allergic rhinitis in the Singapore Chinese population
- Genetic Role in Allergic Rhinitis
- Genetic Factors in Bronchial Asthma