J Korean Med Sci.
2006 Aug;21(4):656-665. 10.3346/jkms.2006.21.4.656.
Genetic Alterations in Primary Gastric Carcinomas Correlated with Clinicopathological Variables by Array Comparative Genomic Hybridization
- Affiliations
-
- 1Department of Clinical Pathology, Chungnam National University Hospital, Daejeon, Korea. shkoo@cnuh.co.kr
- 2Department of Surgery, Chungnam National University Hospital, Daejeon, Korea.
- 3Macrogen, Inc., Seoul, Korea
- KMID: 2157817
- DOI: http://doi.org/10.3346/jkms.2006.21.4.656
Abstract
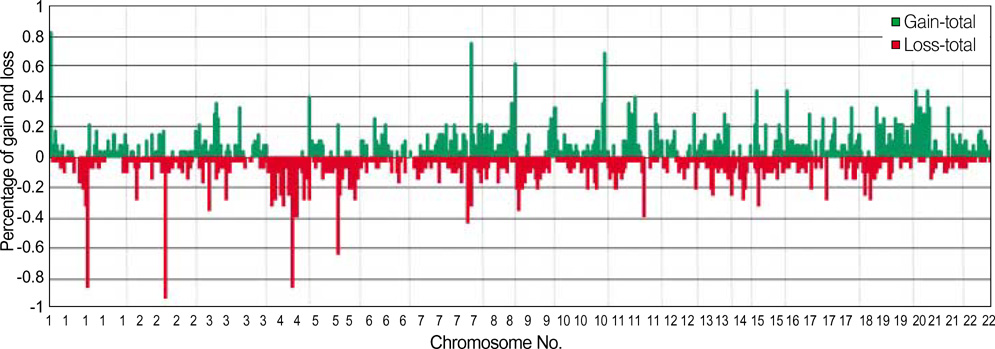
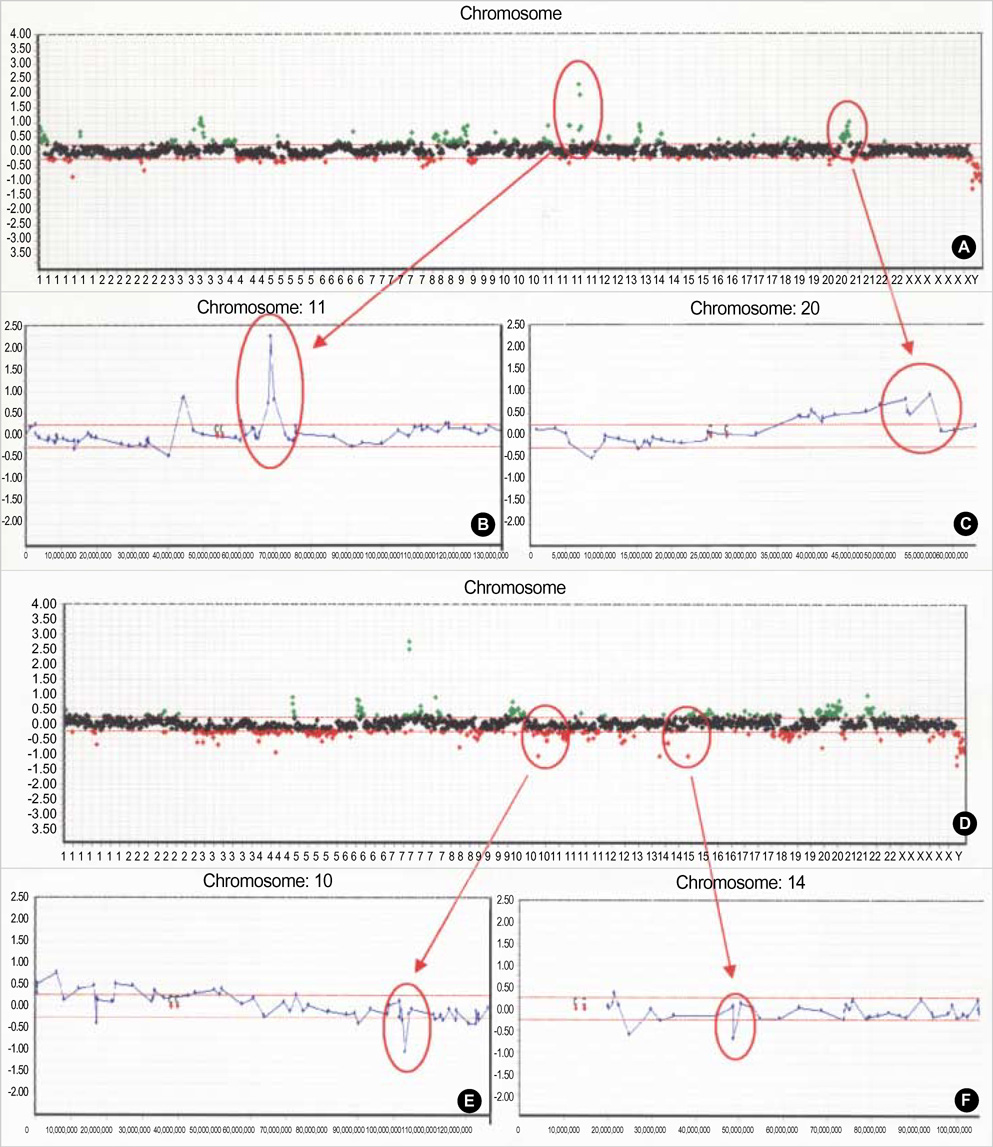
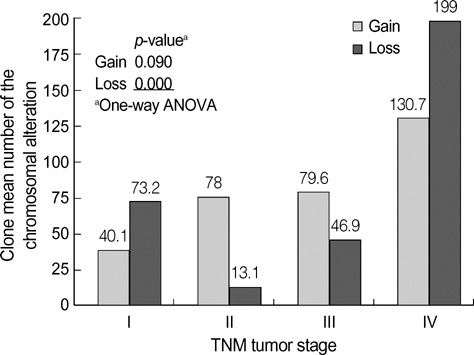
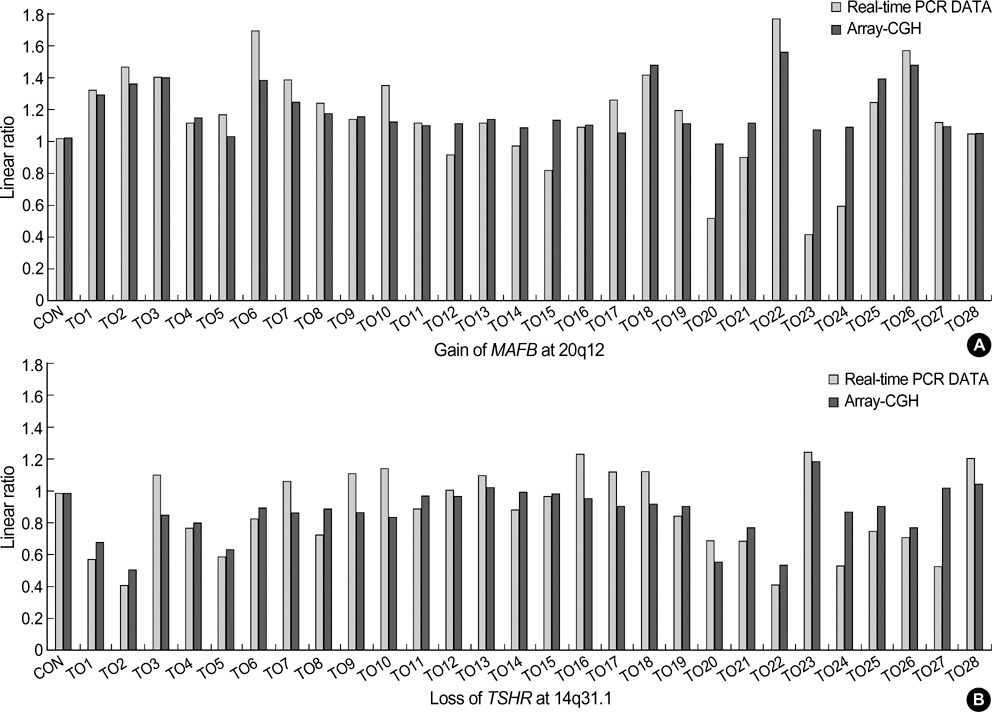
- Genetic alterations have been recognized as an important event in the carcinogenesis of gastric cancer (GC). We conducted high resolution bacterial artificial chromosome array-comparative genomic hybridization, to elucidate in more detail the genomic alterations, and to establish a pattern of DNA copy number changes with distinct clinical variables in GC. Our results showed some correlations between novel amplified or deleted regions and clinical status. Copy-number gains were frequently detected at 1p, 5p, 7q, 8q, 11p, 16p, 20p and 20q, and losses at 1p, 2q, 4q, 5q, 7q, 9p, 14q, and 18q. Losses at 4q23, 9p23, 14q31.1, or 18q21.1 as well as a gain at 20q12 were correlated with tumor-node-metastasis tumor stage. Losses at 9p23 or 14q31.1 were associated with lymph node status. Metastasis was determined to be related to losses at 4q23 or 4q28.2, as well as losses at 4q15.2, 4q21.21, 4q 28.2, or 14q31.1, with differentiation. One of the notable aspects of this study was that the losses at 4q or 14q could be employed in the evaluation of the metastatic status of GC. Our results should provide a potential resource for the molecular cytogenetic events in GC, and should also provide clues in the hunt for genes associated with GC.
Keyword
MeSH Terms
-
Stomach Neoplasms/genetics/*pathology
Reverse Transcriptase Polymerase Chain Reaction/methods
Receptors, Thyrotropin/genetics
Nucleic Acid Hybridization/*methods
Neoplasm Staging
Middle Aged
Male
MafB Transcription Factor/genetics
Lymphatic Metastasis/genetics
Humans
Genome, Human/genetics
Gene Expression Regulation, Neoplastic
Female
Chromosomes, Human, Pair 20/genetics
Chromosomes, Human, Pair 14/genetics
*Chromosome Aberrations
Aged, 80 and over
Aged
Adult
Figure
Reference
-
1. Takada H, Imoto I, Tsuda H, Sonoda I, Ichikura T, Mochizuki H, Okanoue T, Inazawa J. Screening of DNA copy-number aberrations in gastric cancer cell lines by array-based comparative genomic hybridization. Cancer Sci. 2005. 96:100–110.
Article2. Kimura Y, Noguchi T, Kawahara K, Kashima K, Daa T, Yokoyama S. Genetic alterations in 102 primary gastric cancers by comparative genomic hybridization: gain of 20q and loss of 18q are associated with tumor progression. Mod Pathol. 2004. 17:1328–1337.
Article3. Koo SH, Kwon KC, Shin SY, Jeon YM, Park JW, Kim SH, Noh SM. Genetic alterations of gastric cancer: comparative genomic hybridization and fluorescence in situ hybridization studies. Cancer Genet Cytogenet. 2000. 117:97–103.4. Koizumi Y, Tanaka S, Mou R, Koganei H, Kokawa A, Kitamura R, Yamauchi H, Ookubo K, Saito T, Tominaga S, Matsumura K, Shimada H, Tsuchida N, Sekihara H. Changes in DNA copy number in primary gastric carcinomas by comparative genomic hybridization. Clin Cancer Res. 1997. 3:1067–1076.5. Okada K, Sugihara H, Bamba M, Bamba T, Hattori T. Sequential numerical changes of chromosomes 7 and 18 in diffuse-type stomach cancer cell lines: combined comparative genomic hybridization, fluorescence in situ hybridization, and ploidy analyses. Cancer Genet Cytogenet. 2000. 118:99–107.6. Wu MS, Chang MC, Huang SP, Tseng CC, Sheu JC, Lin YW, Shun CT, Lin MT, Lin JT. Correlation of histologic subtypes and replication error phenotype with comparative genomic hybridization in gastric cancer. Genes Chromosomes Cancer. 2001. 30:80–86.
Article7. Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992. 258:818–821.
Article8. Weiss MM, Kuipers EJ, Postma C, Snijders AM, Pinkel D, Meuwissen SG, Albertson D, Meijer GA. Genomic alterations in primary gastric adenocarcinomas correlate with clinicopathological characteristics and survival. Cell Oncol. 2004. 26:307–317.9. Wilhelm M, Veltman JA, Olshen AB, Jain AN, Moore DH, Presti JC Jr, Kovacs G, Waldman FM. Array-based comparative genomic hybridization for the differential diagnosis of renal cell cancer. Cancer Res. 2002. 62:957–960.10. Man TK, Lu XY, Jaeweon K, Perlaky L, Harris CP, Shah S, Ladanyi M, Gorlick R, Lau CC, Rao PH. Genome-wide array comparative genomic hybridization analysis reveals distinct amplifications in osteosarcoma. BMC Cancer. 2004. 4:45.
Article11. Peng DF, Sugihara H, Mukaisho K, Tsubosa Y, Hattori T. Alterations of chromosomal copy number during progression of diffuse-type gastric carcinomas: metaphase-and array-based comparative genomic hybridization analyses of multiple samples from individual tumours. J Pathol. 2003. 201:439–450.12. Kawaguchi K, Honda M, Yamashita T, Shirota Y, Kaneko S. Differential gene alteration among hepatoma cell lines demonstrated by cDNA microarray-based comparative genomic hybridization. Biochem Biophys Res Commun. 2005. 329:370–380.
Article13. Albertson DG, Pinkel D. Genomic microarrays in human genetic disease and cancer. Hum Mol Genet. 2003. 12:145–152.
Article14. Frijters AC, Zhang JZ, van Damme M, Wang GL, Ronald PC, Michelmore RW. Construction of a bacterial artificial chromosome library containing large Eco RI and Hin dIII genomic fragments of lettuce. Theor Appl Genet. 1997. 94:390–399.
Article15. Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, Dairkee SH, Ljung BM, Gray JW, Albertson DG. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998. 20:207–211.
Article16. Shaw CJ, Shaw CA, Yu W, Stankiewicz P, White LD, Beaudet AL, Lupski JR. Comparative genomic hybridisation using a proximal 17p BAC/PAC array detects rearrangements responsible for four genomic disorders. J Med Genet. 2004. 41:113–119.
Article17. Veltman JA, Fridlyand J, Pejavar S, Olshen AB, Korkola JE, DeVries S, Carroll P, Kuo WL, Pinkel D, Albertson D, Cordon-Cardo C, Jain AN, Waldman FM. Array-based comparative genomic hybridization for genome-wide screening of DNA copy number in bladder tumors. Cancer Res. 2003. 63:2872–2880.18. Schneider-Stock R, Boltze C, Lasota J, Miettinen M, Peters B, Pross M, Roessner A. High prognostic value of p16INK4 alterations in gastrointestinal stromal tumors. J Clin Oncol. 2003. 21:1688–1697.
Article19. Kang MJ, Park BJ, Byun DS, Park JI, Kim HJ, Park JH, Chi SG. Loss of imprinting and elevated expression of wild-type p73 in human gastric adenocarcinoma. Clin Cancer Res. 2000. 6:1767–1771.20. Chan WY, Liu Y, Li CY, Ng EK, Chow JH, Li KK, Chung SC. Recurrent genomic aberrations in gastric carcinomas associated with Helicobacter pylori and Epstein-Barr virus. Diagn Mol Pathol. 2002. 11:127–134.
Article21. Derre J, Lagace R, Terrier P, Sastre X, Aurias A. Consistent DNA losses on the short arm of chromosome 1 in a series of malignant gastrointestinal stromal tumors. Cancer Genet Cytogent. 2001. 127:30–33.22. O'Leary T, Ernst S, Przygodzki R, Emory T, Sobin L. Loss of heterozygosity at 1p36 predicts poor prognosis in gastrointestinal stromal/smooth muscle tumors. Lab Invest. 1999. 79:1461–1467.23. Hodgson JG, Chin K, Collins C, Gray JW. Genome amplification of chromosome 20 in breast cancer. Breast Cancer Res Treat. 2003. 78:337–345.
Article24. Wang Q, Wang B, Guan X, Gao H, Cheng H, Zhang Q, Huang C, Li P. Analysis of comparative genomic hybridization and loss of heterozygosity in 43 primary gastric carcinomas. Chin Med J. 2003. 116:517–523.25. Rumpel CA, Powell SM. Mapping of genetic deletions on the long arm of chromosome 4 in human esophageal adenocarcinomas. Am J Pathol. 1999. 154:1329–1334.
Article26. Yan WS, Song LY, Wei WD, Li A, Liang QW, Liu JH, Fang Y. Chromosomal imbalance in primary lung squamous cell carcinoma and their relationship with smoking. Ai Zheng. 2005. 24:47–52.27. Kallio JP, Mahlamaki EH, Helin H, Karhu R, Kellokumpu-Lehtinen P, Tammela TL. Chromosomal gains and losses detected by comparative genomic hybridization and proliferation activity in renal cell carcinoma. Scand J Urol Nephrol. 2004. 38:225–230.28. Jones C, Ford E, Gillett C, Ryder K, Merrett S, Reis-Filho JS, Fulford LG, Hanby A, Lakhani SR. Molecular cytogenetic identification of subgroups of grade III invasive ductal breast carcinomas with different clinical outcomes. Clin Cancer Res. 2004. 10:5988–5997.
Article29. Van Dekken H, Geelen E, Dinjens WN, Wijnhoven BP, Tilanus HW, Tanke HJ, Rosenberg C. Comparative genomic hybridization of cancer of the gastroesophageal junction: deletion of 14Q31-32. 1 discriminates between esophageal (Barrett's) and gastric cardia adenocarcinomas. Cancer Res. 1999. 59:748–752.30. Koo SH, Jeong TE, Kang J, Kwon KC, Park JW, Noh SM. Prognostic implications for gastric carcinoma based on loss of heterozygosity genotypes correlation with clinicopathologic variables. Cancer Genet Cytogenet. 2004. 153:26–31.31. Nishizuka S, Tamura G, Terashima M, Satodate R. Loss of heterozygosity during the development and progression of differentiated adenocarcinoma of the stomach. J Pathol. 2000. 185:38–43.
Article32. Choi SW, Choi JR, Chung YJ, Kim KM, Rhyu MG. Prognostic implications of microsatellite genotypes in gastric carcinoma. Int J Cancer. 2000. 89:378–383.
Article33. Kim NG, Kim JJ, Ahn JY, Seong CM, Noh SH, Kim CB, Min JS, Kim H. Putative chromosomal deletions on 9p, 9q and 22q occur preferentially in malignant gastrointestinal stromal tumors. Int J Cancer. 2000. 85:633–638.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Array-based Comparative Genomic Hybridization and Its Application to Cancer Genomes and Human Genetics
- Chromosomal gains and losses in primary ovarian carcinomas by comparative genomic hybridization
- Genetic Alterations in Gastric Carcinomas and Adjacent Mucosa Detected by Comparative Genomic Hybridization (CGH)
- Current Status and Future Clinical Applications of Array.based Comparative Genomic Hybridization
- Next generation sequencing and array-based comparative genomic hybridization for molecular diagnosis of pediatric endocrine disorders