J Bacteriol Virol.
2007 Jun;37(2):79-89. 10.4167/jbv.2007.37.2.79.
Genetic Diversity of Stenotrophomonas maltophilia Isolated from Clinical Specimens
- Affiliations
-
- 1Department of Microbiology, Kyungpook National University School of Medicine, Daegu 700-422, Korea. yclee@knu.ac.kr
- KMID: 2055042
- DOI: http://doi.org/10.4167/jbv.2007.37.2.79
Abstract
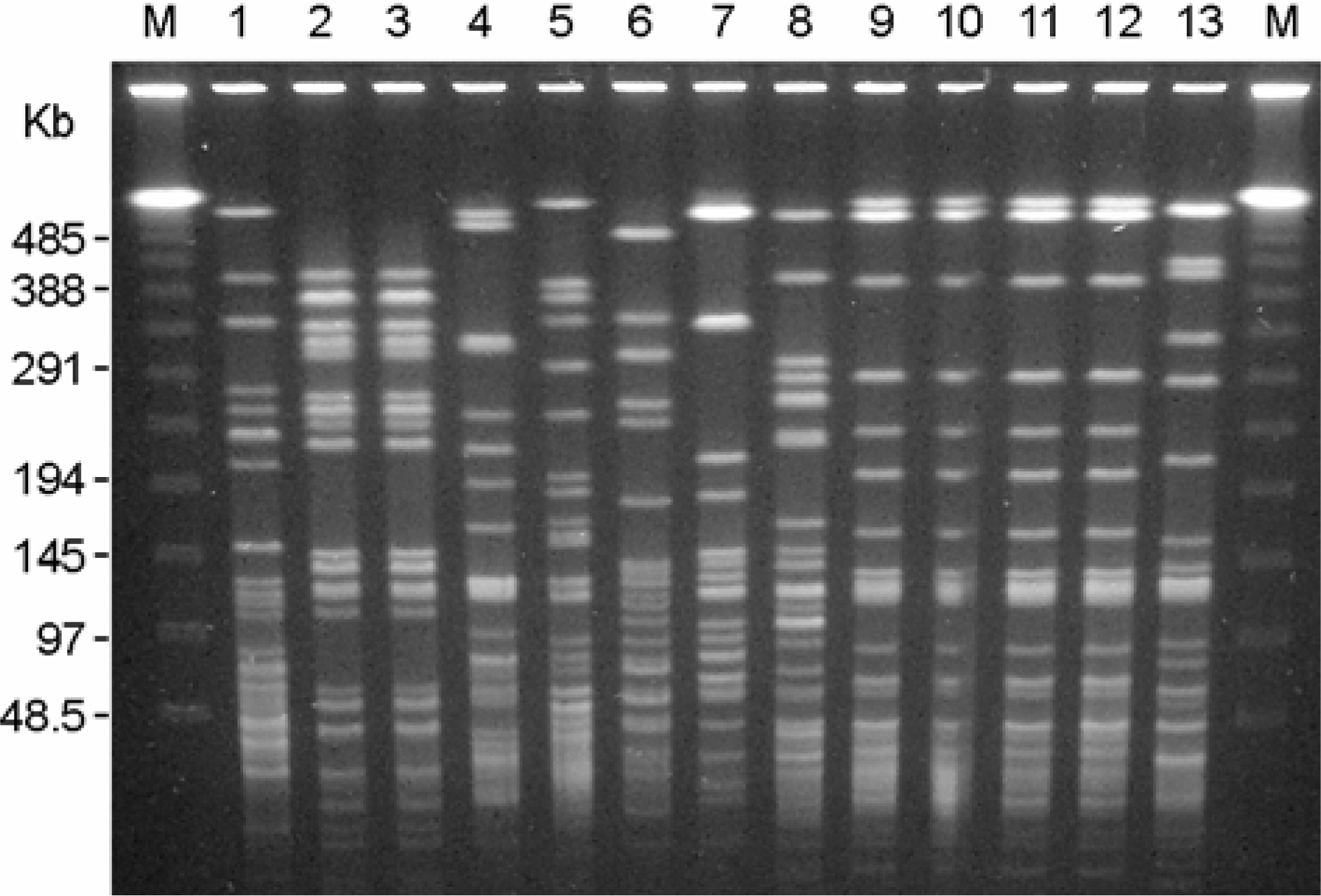
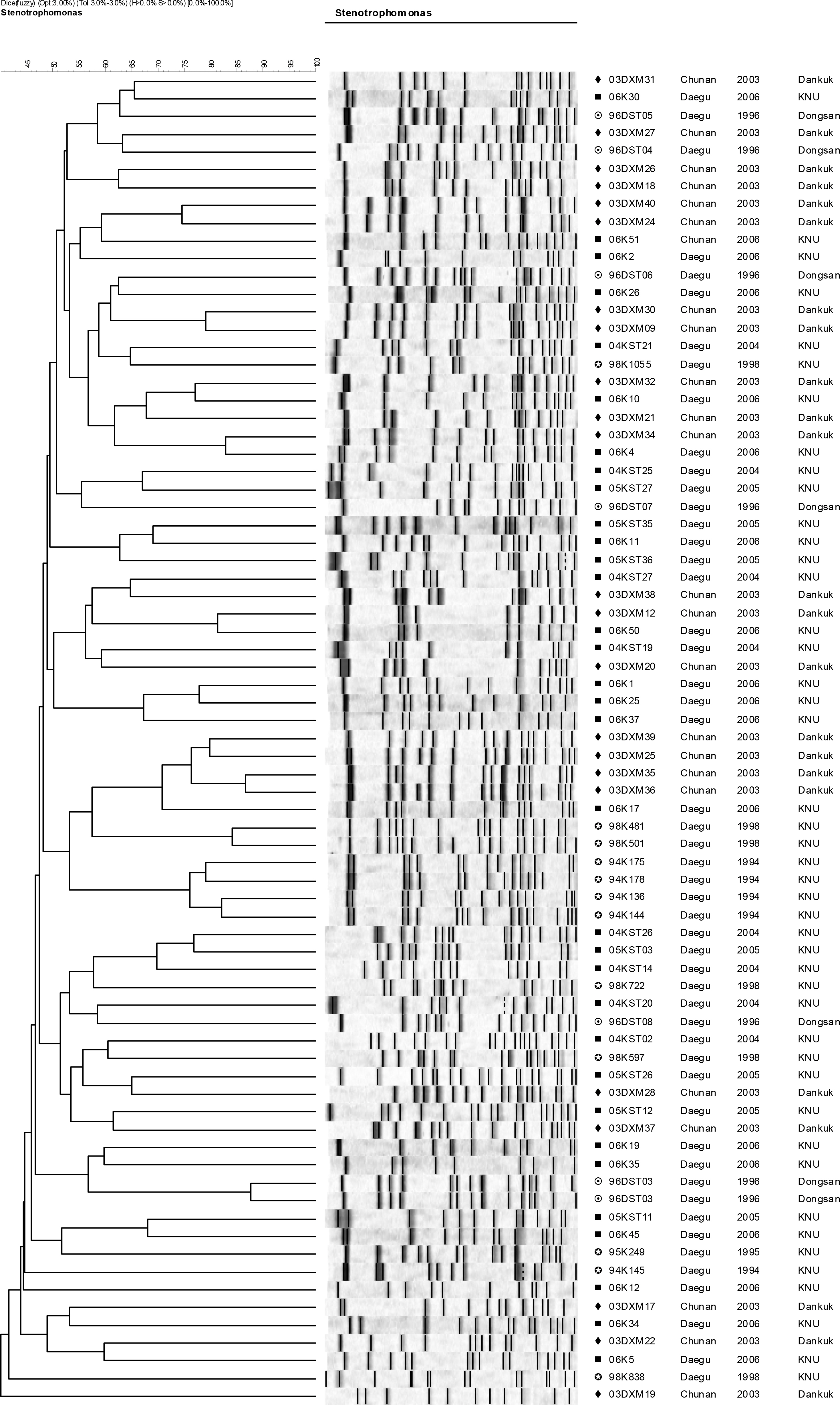
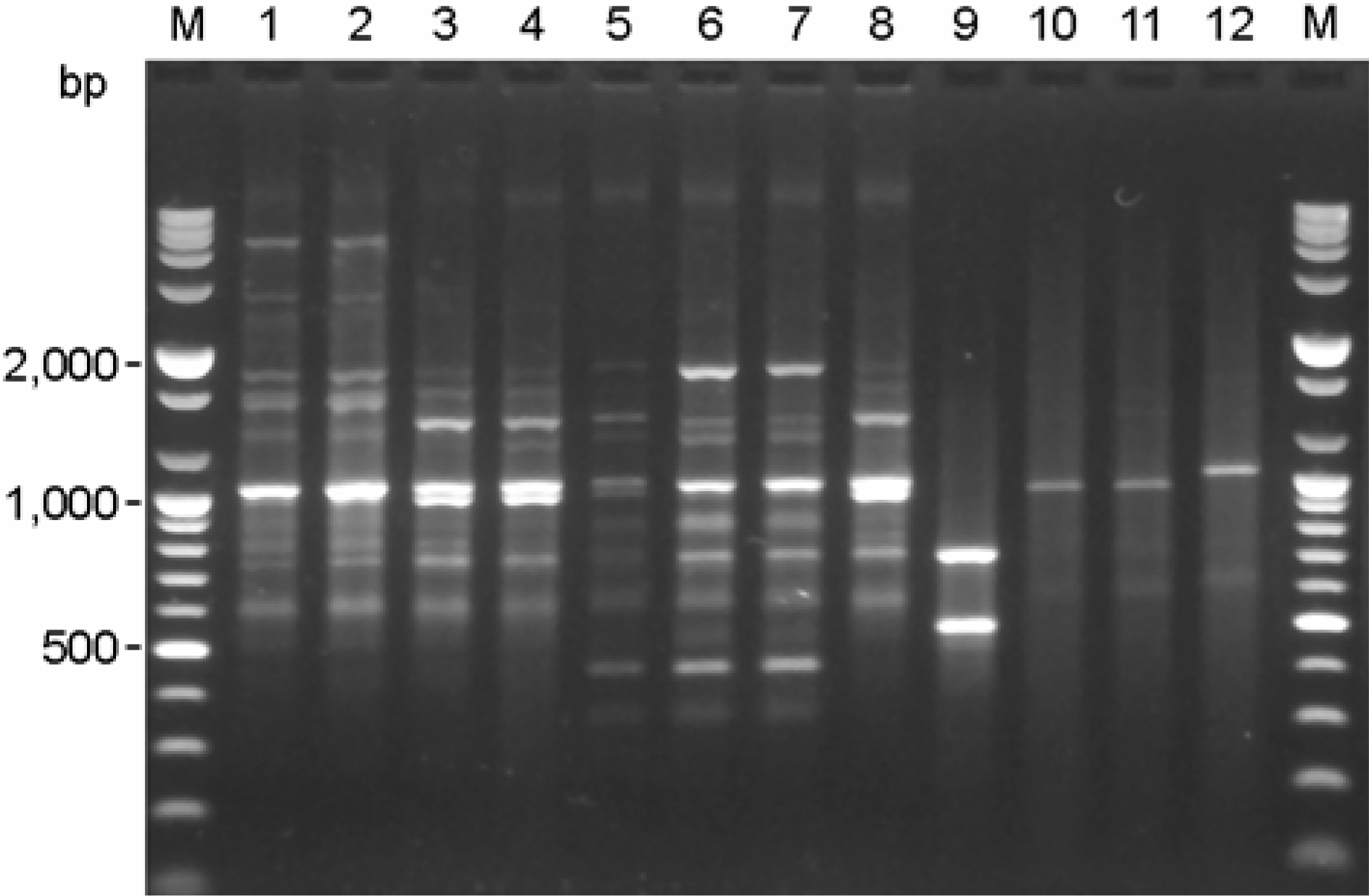
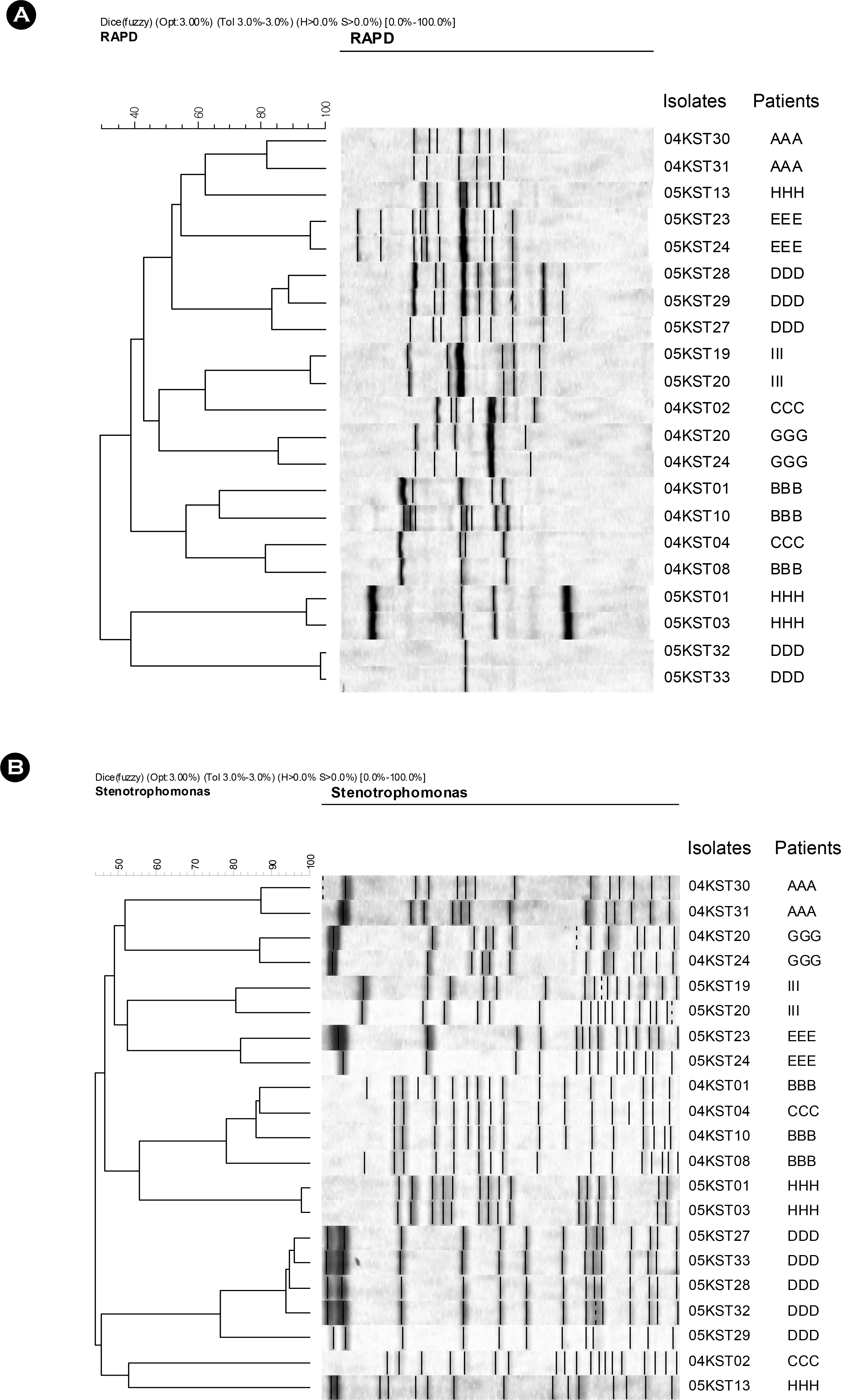
- Stenotrophomonas maltophilia is a multi-drug resistant pathogen that has been isolated with increasing frequency from the hospitalized patients. A total of 202 S. maltophilia was isolated from three university hospitals and analysed by molecular typing for an epidemiologic investigation. All isolates were tested by antimicrobial susceptibility, random amplified polymorphic DNA (RAPD) analysis, and pulsed-field gel electrophoresis (PFGE). The RAPD and PFGE patterns were recorded and analysed by the unweighted-pair group method with arithmetic average method. Two or more isolates were considered to be clonally related if their PFGE pattern exhibited > or =80% similarity. Trimethoprim/ sulfamethoxazole and ciprofloxacin were the most active antimicrobial agents tested. The majority of the isolates found to be genetically unrelated by PFGE. The genetically related isolates were recovered from the same patient. The result demonstrates a high genetic diversity of S. maltophilia isolates from clinical specimens. The clonal diversity of S. maltophilia from the hospitalized patients is partly due to the strains originated from the hospital environments, but not horizontal transfer between the patients
MeSH Terms
Figure
Reference
-
References
1). 설성용, 장경수, 정웅기, 조응래, 김능희, 유학선, 이유 철. 병원 재료에서 분리한 Stenotrophomonas maltophilia 의 항균제 내성 및 분자역학적 특성. 대한미생물학회지 Stenotrophomonas maltophilia. 35:239–250. 2000.2). Babalova M, Blahova J, Lesicka-Hupkova M, Krcmery V, Kubonova K. Transfer of ceftazydime and aztreonam resistance from nosocomial strains of Xanthomonas (Stenotrophomonas) maltiphilia to a recipient strain of Pseudomonas aeruginosa ML-1008. Eur J Clin Microbiol Infect Dis. 14:925–926. 1995.3). Berg G, Roskot N, Smalla K. Genotypic and phenotypic relationships between clinical and environmental isolates of Stenotrophomonas maltophilia. J Clin Microbiol. 37:3594–3600. 1999.4). Bingen EH, Denamur E, Lambert-Zechovsky NY, Bourdois A, Mariani-Kurkdjian P, Cezard JP, Navarro J, Elion J. DNA restriction fragment lengh polymorphism differentiates crossed from independent infections in nosocomial Xanthomonas maltiphilia bacteremia. J Clin Microbiol. 29:1348–1350. 1991.5). Chang LL, Chen HF, Chang CY, Lee TM, Wu WJ. Contribution of integrons, and SmeABC and SmeDEF efflux pumps to multidrug resistance in clinical isolates of Stenotrophomonas maltophilia. J Antimicrob Chemother. 53:518–521. 2004.6). Chang N, Chui L. A standardized protocol for the rapid preparation of bacterial DNA for pulsed-field gel electrophoresis. Diagn Microbiol Infect Dis. 31:275–279. 1998.
Article7). Chatelut M, Dournes JL, Chabanon G, Marty N. Epidemiological typing of Stenotrophomonas (Xanthomonas) maltophilia by PCR. J Clin Microbiol. 33:912–914. 1995.8). Chow AW, Wong J, Bartlett KH. Synergistic interactions of ciprofloxacin and extended-spectrum beta-lactams or aminoglycosides against multiple drug-resistant Pseudomonas maltophilia. Antimicrob Agents Chemother. 32:782–784. 1988.9). Denton M, Kerr KG. Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin Microbiol Rev. 11:57–80. 1998.10). Denton M, Todd NJ, Kerr KG, Hawkey PM, Littlewood JM. Molecular epidemiology of Stenotrophomonas maltophilia isolated from clinical specimens from patients with cystic fibrosis and associated environmental samples. J Clin Microbiol. 36:1953–1958. 1998.11). Gales AC, Jones RN, Forward KR, Linares J, Sader HS, Verhoef J. Emerging importance of multidrug-resistant Acinetobacter species and Stenotrophomonas maltophilia as pathogens in seriously ill patients: geographic patterns, epidemiological features, and trends in the SENTRY Antimicrobial Surveillance Program (1997∼1999). Clin Infect Dis. 32:S104–S113. 2001.12). Gerner-Smidt P, Bruun B, Arpi M, Schmidt J. Diversity of nosocomial Xanthomonas maltophilia (Stenotrophomonas maltophilia) as determined by ribotyping. Eur J Clin Microbiol Infect Dis. 14:137–140. 1995.13). HPA. Pseudomonas spp and Stenotrophomonas maltophilia bacteraemia: England, Wales, and Northern Ireland 2003. CDR Weekly Volume. 14:1–7. No 43,. 2004.14). Jones RN, Sader HS, Beach ML. Contemporary in vitro spectrum of activity summary for antimicrobial agents tested against 18569 strains non-fermentative Gram-negative bacilli isolated in the SENTRY Antimicrobial Surveillance Program (1997∼2001). Int J Antimicrob Agents. 22:551–556. 2003.
Article15). Sader HS, Jones RN. Antimicrobial susceptibility of uncommonly isolated non-enteric Gram-negative bacilli. Int J Antimicrob Agents. 25:95–109. 2005.
Article16). Kim JM, Park ES, Jeong JS, Kim KM, Kim JM, Oh HS, Yoon SW, Chang HS, Chang KH, Lee SI, Lee MS, Song JH, Kang MW, Park SC, Choe KW, Pai CH. Multicenter surveillance study for nosocomial infections in major hospitals in Korea. Nosocomial Infection Surveillance Committee of the Korean Society for Nosocomial Infection Control. Am J Infect Control. 28:454–458. 2000.17). Kim JH, Kim SW, Kang HR, Bae GB, Park JH, Nam EJ, Kang YM, Lee JM, Kim NS. Two episodes of Stenotrophomonas maltophilia endocarditis of prosthetic mitral valve: report of a case and review of the literature. J Korean Med Sci. 17:263–265. 2002.18). Laing FP, Ramotar K, Read RR, Alfieri N, Kureishi A, Henderson EA, Louie TJ. Molecular epidemiology of Xanthomonas maltophilia colonization and infection in the hospital environment. J Clin Microbiol. 33:513–518. 1995.19). Meyer E, Schwab F, Gastmeier P, Ruden H, Daschner FD. Is the prevalence of Stenotrophomonas maltophilia isolation and nosocomial infection increasing in intensive care units? Eur J Clin Microbiol Infect Dis. 25:711–714. 2006.20). Morrison AJ Jr, Hoffmann KK, Wenzel RP. Associated mortality and clinical characteristics of nosocomial Pseudomonas maltophilia in a university hospital. J Clin Microbiol. 24:52–55. 1986.21). National Committee for Clinical Labratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, National Committee for Clinical Laboratory Standards. Approved document M7-A6, Wayne, Pa. 2003.22). Pankuch GA, Jacobs MR, Appelbaum PC. Susceptibilities of 123 Xanthomonas maltophilia strains to clinafloxacin, PD 131628, PD 138312, PD 140248, ciprofloxacin, and ofloxacin. Antimicrob Agents Chemother. 38:369–370. 1994.23). Poulos CD, Matsumura SO, Willey BM, Low DE, McGeer A. In vitro activities of antimicrobial combinations against Stenotrophomonas (Xanthomonas) maltophilia. Antimicrob Agents Chemother. 39:2220–2223. 1995.24). Schmitz FJ, Sadurski R, Verhoef J, Milatovic D, Fluit AC. Typing of 154 clinical isolates of Stenotrophomonas maltophilia by pulsed-field gel electrophoresis and determination of the in vitro susceptibilities of these strains to 28 antibiotics. J Antimicrob Chemother. 45:921–923. 2000.25). Senol E. Stenotrophomonas maltophilia: the significance and role as a nosocomial pathogen. J Hosp Infect. 57:1–7. 2004.26). Valdezate S, Vindel A, Martin-Davila P, Del Saz BS, Baquero F, Canton R. High genetic diversity among Stenotrophomonas maltophilia strains despite their originating at a single hospital. J Clin Microbiol. 42:693–699. 2004.27). VanCouwenberghe CJ, Cohen SH, Tang YJ, Gumerlock PH, Silva J Jr. Genomic fingerprinting of epidemic and endemic strains of Stenotrophomonas maltophilia (formerly Xanthomonas maltophilia) by arbitrarily primed PCR. J Clin Microbiol. 33:1289–1291. 1995.28). Whitby PW, Carter KB, Burns JL, Royall JA, LiPuma JJ, Stull TL. Identification and detection of Stenotrophomonas maltophilia by rRNA-directed PCR. J Clin Microbiol. 38:4305–4309. 2000.29). Yao JD, Conly JM, Krajden M. Molecular typing of Stenotrophomonas (Xanthomonas) maltophilia by DNA macrorestriction analysis and random amplified polymorphic DNA analysis. J Clin Microbiol. 33:2195–2198. 1995.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Case of Indophenol Oxidase-positive Stenotrophomonas maltophilia Isolated from Urine in a Patient with Acute Lymphoblastic Leukemia
- Localized Cutaneous Infection due to Stenotrophomonas maltophilia in Immunocompetent Patient
- A Case of Corneal Ulcer Caused by Combined Infection of Stenotrophomonas Maltophilia and Aspergillus Fumigatus
- A Case of Stenotrophomonas Maltophilia Endophthalmitis after Cataract Operation
- Clinical Examination of Stenotrophomonas maltophilia Infection in Patient with Pyoderma Gangrenosum