Tuberc Respir Dis.
2012 Sep;73(3):137-142. 10.4046/trd.2012.73.3.137.
Year-in-Review of Lung Cancer
- Affiliations
-
- 1Department of Internal Medicine, Konyang University Hospital, Konyang University College of Medicine, Daejeon, Korea. sk1609@hanmail.net
- KMID: 1842908
- DOI: http://doi.org/10.4046/trd.2012.73.3.137
Abstract
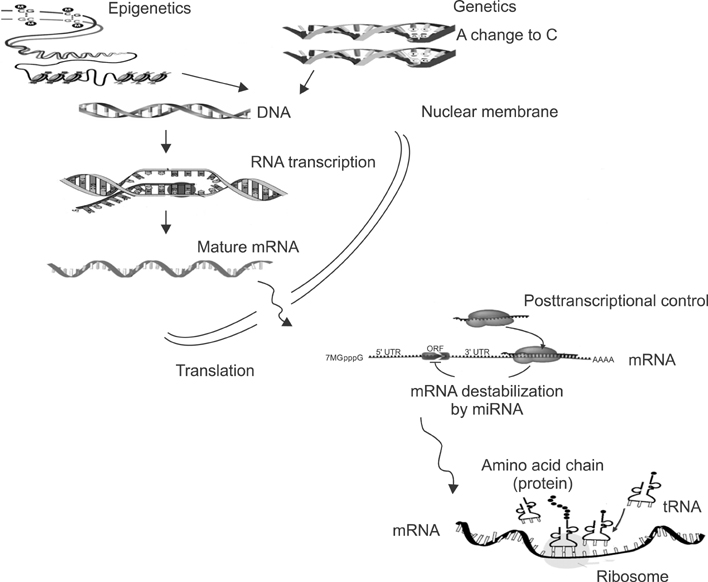
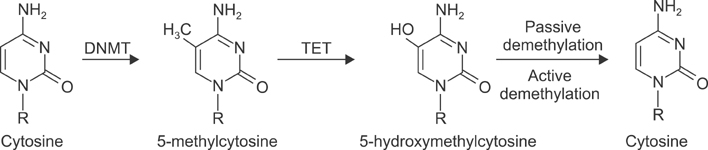
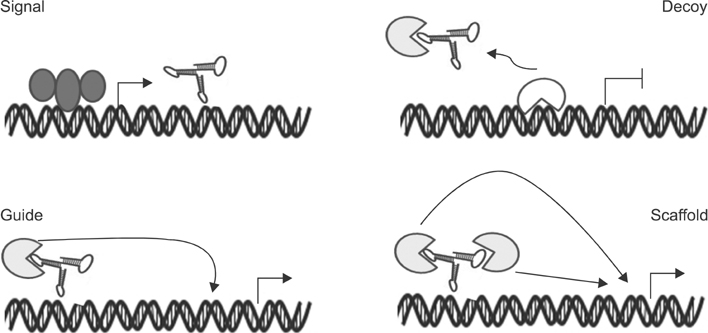
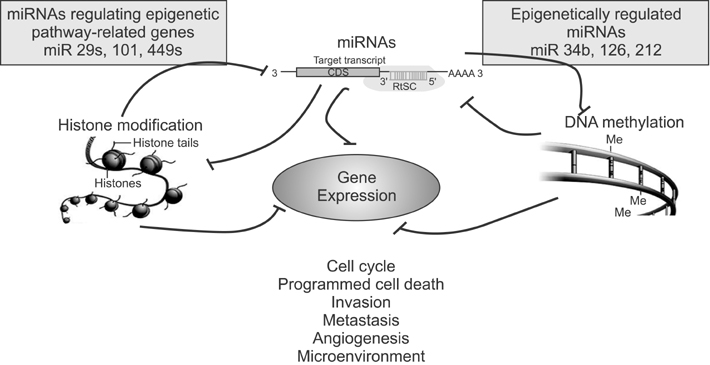
- In the last several years, we have made slow but steady progress in understanding molecular biology of lung cancer. This review is focused on advances in understanding the biology of lung cancer that have led to proof of concept studies on new therapeutic approaches. The three selected topics include genetics, epigenetics and non-coding RNA. This new information represents progress in the integration of molecular mechanisms that to identify more effective ways to target lung cancer.
Figure
Reference
-
1. Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010. 60:277–300.2. Spiro SG, Silvestri GA. One hundred years of lung cancer. Am J Respir Crit Care Med. 2005. 172:523–529.3. Kulesza P, Ramchandran K, Patel JD. Emerging concepts in the pathology and molecular biology of advanced non-small cell lung cancer. Am J Clin Pathol. 2011. 136:228–238.4. Govindan R. Summary of presentation from the targeted therapy in lung cancer meeting. J Thorac Oncol. 2011. 6:11 Suppl 4. S1757.5. Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010. 11:685–696.6. Lipson D, Capelletti M, Yelensky R, Otto G, Parker A, Jarosz M, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012. 18:382–384.7. Ju YS, Lee WC, Shin JY, Lee S, Bleazard T, Won JK, et al. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res. 2012. 22:436–445.8. Jung Y, Kim P, Jung Y, Keum J, Kim SN, Choi YS, et al. Discovery of ALK-PTPN3 gene fusion from human non-small cell lung carcinoma cell line using next generation RNA sequencing. Genes Chromosomes Cancer. 2012. 51:590–597.9. Togashi Y, Soda M, Sakata S, Sugawara E, Hatano S, Asaka R, et al. KLC1-ALK: a novel fusion in lung cancer identified using a formalin-fixed paraffin-embedded tissue only. PLoS One. 2012. 7:e31323.10. Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002. 3:415–428.11. Son JW, Kim YJ, Cho HM, Lee SY, Lee SM, Kang JK, et al. Promoter hypermethylation of the CFTR gene and clinical/pathological features associated with non-small cell lung cancer. Respirology. 2011. 16:1203–1209.12. Gery S, Komatsu N, Kawamata N, Miller CW, Desmond J, Virk RK, et al. Epigenetic silencing of the candidate tumor suppressor gene Per1 in non-small cell lung cancer. Clin Cancer Res. 2007. 13:1399–1404.13. Hamilton JP. Epigenetics: principles and practice. Dig Dis. 2011. 29:130–135.14. Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2012. 13:28–35.15. Jin SG, Jiang Y, Qiu R, Rauch TA, Wang Y, Schackert G, et al. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res. 2011. 71:7360–7365.16. Kudo Y, Tateishi K, Yamamoto K, Yamamoto S, Asaoka Y, Ijichi H, et al. Loss of 5-hydroxymethylcytosine is accompanied with malignant cellular transformation. Cancer Sci. 2012. 103:670–676.17. Eddy SR. Non-coding RNA genes and the modern RNA world. Nat Rev Genet. 2001. 2:919–929.18. ENCODE Project Consortium. Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007. 447:799–816.19. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011. 43:904–914.20. Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011. 10:38.21. Schmidt LH, Spieker T, Koschmieder S, Schäffers S, Humberg J, Jungen D, et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011. 6:1984–1992.22. Sato F, Tsuchiya S, Meltzer SJ, Shimizu K. MicroRNAs and epigenetics. FEBS J. 2011. 278:1598–1609.23. Watanabe K, Emoto N, Hamano E, Sunohara M, Kawakami M, Kage H, et al. Genome structure-based screening identified epigenetically silenced microRNA associated with invasiveness in non-small-cell lung cancer. Int J Cancer. 2012. 130:2580–2590.24. Incoronato M, Urso L, Portela A, Laukkanen MO, Soini Y, Quintavalle C, et al. Epigenetic regulation of miR-212 expression in lung cancer. PLoS One. 2011. 6:e27722.25. Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setién F, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007. 67:1424–1429.26. Zhang JG, Guo JF, Liu DL, Liu Q, Wang JJ. MicroRNA-101 exerts tumor-suppressive functions in non-small cell lung cancer through directly targeting enhancer of zeste homolog 2. J Thorac Oncol. 2011. 6:671–678.27. Jeon HS, Lee SY, Lee EJ, Yun SC, Cha EJ, Choi E, et al. Combining microRNA-449a/b with a HDAC inhibitor has a synergistic effect on growth arrest in lung cancer. Lung Cancer. 2012. 76:171–176.28. Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007. 104:15805–15810.